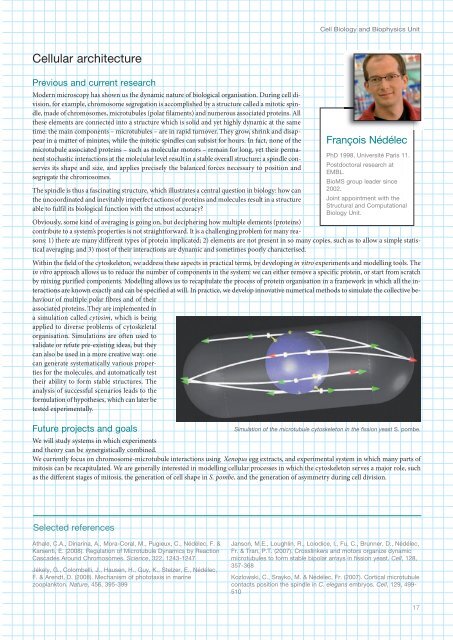
Cell Biology and Biophysics UnitCellular architecturePrevious and current researchModern microscopy has shown us the dynamic nature of biological organisation. During cell division,for example, chromosome segregation is accomplished by a structure called a mitotic spindle,made of chromosomes, microtubules (polar filaments) and numerous associated proteins. Allthese elements are connected into a structure which is solid and yet highly dynamic at the sametime: the main components – microtubules – are in rapid turnover. They grow, shrink and disappearin a matter of minutes, while the mitotic spindles can subsist for hours. In fact, none of themicrotubule associated proteins – such as molecular motors – remain for long, yet their permanentstochastic interactions at the molecular level result in a stable overall structure: a spindle conservesits shape and size, and applies precisely the balanced forces necessary to position andsegregate the chromosomes.The spindle is thus a fascinating structure, which illustrates a central question in biology: how canthe uncoordinated and inevitably imperfect actions of proteins and molecules result in a structureable to fulfil its biological function with the utmost accuracy?Obviously, some kind of averaging is going on, but deciphering how multiple elements (proteins)contribute to a system’s properties is not straightforward. It is a challenging problem for many reasons:1) there are many different types of protein implicated; 2) elements are not present in so many copies, such as to allow a simple statisticalaveraging; and 3) most of their interactions are dynamic and sometimes poorly characterised.Within the field of the cytoskeleton, we address these aspects in practical terms, by developing in vitro experiments and modelling tools. Thein vitro approach allows us to reduce the number of components in the system: we can either remove a specific protein, or start from scratchby mixing purified components. Modelling allows us to recapitulate the process of protein organisation in a framework in which all the interactionsare known exactly and can be specified at will. In practice, we develop innovative numerical methods to simulate the collective behaviourof multiple polar fibres and of theirassociated proteins. They are implemented ina simulation called cytosim, which is beingapplied to diverse problems of cytoskeletalorganisation. Simulations are often used tovalidate or refute pre-existing ideas, but theycan also be used in a more creative way: onecan generate systematically various propertiesfor the molecules, and automatically testtheir ability to form stable structures. Theanalysis of successful scenarios leads to theformulation of hypotheses, which can later betested experimentally.Future projects and goalsFrançois NédélecPhD 1998, Université Paris 11.Postdoctoral research at<strong>EMBL</strong>.BioMS group leader since2002.Joint appointment with theStructural and ComputationalBiology Unit.Simulation of the microtubule cytoskeleton in the fission yeast S. pombe.We will study systems in which experimentsand theory can be synergistically combined.We currently focus on chromosome-microtubule interactions using Xenopus egg extracts, and experimental system in which many parts ofmitosis can be recapitulated. We are generally interested in modelling cellular processes in which the cytoskeleton serves a major role, suchas the different stages of mitosis, the generation of cell shape in S. pombe, and the generation of asymmetry during cell division.Selected referencesAthale, C.A., Dinarina, A., Mora-Coral, M., Pugieux, C., Nédélec, F. &Karsenti, E. (2008). Regulation of Microtubule Dynamics by ReactionCascades Around Chromosomes. Science, 322, 123-127Jékely, G., Colombelli, J., Hausen, H., Guy, K., Stelzer, E., Nédélec,F. & Arendt, D. (2008). Mechanism of phototaxis in marinezooplankton. Nature, 56, 395-399Janson, M.E., Loughlin, R., Loiodice, I., Fu, C., Brunner, D., Nédélec,Fr. & Tran, P.T. (2007). Crosslinkers and motors organize dynamicmicrotubules to form stable bipolar arrays in fission yeast. Cell, 128,357-368Kozlowski, C., Srayko, M. & Nédélec, Fr. (2007). Cortical microtubulecontacts position the spindle in C. elegans embryos. Cell, 129, 99-51017
<strong>EMBL</strong> Research at a Glance 2009RainerPepperkokPhD 1992, University ofKaiserslautern.Postdoctoral work atUniversity of Geneva.Lab head at the ImperialCancer Research Fund,London.Team leader at <strong>EMBL</strong> since1998.Membrane traffic in the early secretory pathwayPrevious and current researchTransport between the endoplasmic reticulum (ER) and the Golgi complex in mammalian cells involvesat least four basic steps (see figure): 1) biogenesis of membrane bounded transport carriersat specialised domains (ER-exit sites) of the ER; 2) microtubule mediated transport of thecarriers to the Golgi complex; 3) docking and fusion of the carriers with the Golgi complex; and4) recycling of the transport machinery back to the ER. To warrant both the specificity of deliveryof cargo and the maintenance of the unique protein and lipid compositions of the organellesinvolved, these four steps must be tightly regulated and coordinated at the molecular level.The specific questions we are presently addressing in this context are: 1) what are the mechanismsunderlying the regulation of ER-exit sites biogenesis and function; 2) how are ER exit and microtubulemediated ER to Golgi transport coupled at the molecular level; 3) what are the mechanismsof Golgi biogenesis; and 4) which are themolecules regulating recycling of Golgiresident proteins to the ER.To investigate this, we develop computerautomated light microscopy approachesto directly visualise and quantify in livingcells the kinetics of secretory and organellemarkers simultaneously withvesicular coat molecules (COPI and COPII) and their regulators. We also use fluorescencerecovery after photobleaching (FRAP) and fluorescence resonance energytransfer measurements (FRET), together with mathematical modelling of the data inorder to understand the mechanistic of the temporal and spatial regulation of themolecular interactions involved. Our combined data suggest that secretory cargo,lipids and the microtubule motor associated dynactin complex play a critical role inthe stabilisation of the COPII vesicular coat complex to provide the time that is necessaryfor cargo selection and concentration at ER exit sites. In order to investigatethe mechanisms of Golgi biogenesis we have developed an approach, in which we removeby laser nanosurgery the entire Golgi complex from living cells and subsequentlyanalyse the “Golgi-less” karyoplast by time-lapse and electron microscopy.With this approach we could show that Golgi biogenesis in mammalian cells occursde novo from ER derived membranes.In order to identify putative molecules involved in this de novo Golgi biogenesis, wehave developed and applied functional assays to assess the effect of knock-ins bycDNA over-expression and knockdowns by RNAi, on processes such as constitutiveprotein transport, Golgi integrity and function of vesicular coat complexes. Toachieve the throughput that such genome-wide analyses require we have developeda fully automated high content screening microscopy platform including sample preparation, image acquisition and automated analysis of complexcellular phenotypes. We have applied this technology to genome-wide siRNA screens to identify and characterise comprehensively thegenes and their underlying functional networks involved in secretory membrane traffic and Golgi integrity.Future projects and goalsThe four steps involved in ER to Golgi transport inmammalian cells. (I): Biogenesis of COPII coatedvesicles occurs at specialised ER exit sites of the ER.(II): COPII vesicles homotypically fuse to form largervesicular tubular transport carriers (VTCs) that aretransported to the Golgi complex along microtubules.(III): VTCs arrive at the Golgi complex and fuse to it todeliver their cargo. (IV): Transport machinery andmisrouted proteins are return back to the ER by adistinct class of carriers.We will study the novel proteins, which we revealed in our screens to be involved in the early secretory pathway, in further detail at the systemslevel. An important question in this context will be if and how they participate in the temporal and spatial organisation of ER-exit sitesand their function, and the biogenesis of the Golgi complex.Selected referencesSimpson, J.C., Cetin, C., Erfle, H., Joggerst, B., Liebel, U., Ellenberg,J. & Pepperkok, R. (2007). An RNAi screening platform to identifysecretion machinery in mammalian cells. J. Biotechnol., 129, 352-365Runz, H., Miura, K., Weiss, M. & Pepperkok, R. (2006). Sterolsregulate ER-export dynamics of secretory cargo protein ts-O5-G.EMBO J., 25, 2953-2965Forster, R., Weiss, M., Zimmermann, T., Reynaud, E.G., Verissimo,F., Stephens, D.J. & Pepperkok, R. (2006). Secretory cargo regulatesthe turnover of COPII subunits at single ER exit sites. Curr. Biol., 16,173-179Watson, P., Forster, R., Palmer, K.J., Pepperkok, R. & Stephens, D.J.(2005). Coupling of ER exit to microtubules through direct interactionof COPII with dynactin. Nat. Cell Biol., 7, 8-5518