International Society of Blood Transfusion Working Party on red cell ...
International Society of Blood Transfusion Working Party on red cell ...
International Society of Blood Transfusion Working Party on red cell ...
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
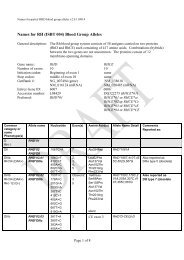
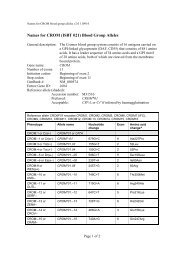
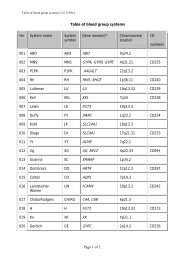
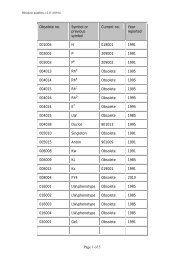
REPORTVox Sanguinis (2011) 101, 77–82ª 2011 The Author(s)Vox Sanguinis ª 2011 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>DOI: 10.1111/j.1423-0410.2010.01462.x<str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> <str<strong>on</strong>g>Working</str<strong>on</strong>g> <str<strong>on</strong>g>Party</str<strong>on</strong>g> <strong>on</strong><strong>red</strong> <strong>cell</strong> immunogenetics and blood group terminology: BerlinreportJ. R. Storry, 1 L. Castilho, 2 G. Daniels, 3 W. A. Flegel, 4 G. Garratty, 5 C. L. Francis, 6 J. M. Moulds, 7 J. J. Moulds, 7 M. L. Olss<strong>on</strong>, 8J. Poole, 3 M. E. Reid, 6 P. Rouger, 9 E. van der Schoot, 10 M. Scott, 3 E. Smart, 11 Y. Tani, 12 L.-C. Yu, 13 S. Wendel, 14 C. Westh<str<strong>on</strong>g>of</str<strong>on</strong>g>f, 6V. Yahalom 15 & T. Zelinski 161 Clinical Immunology and <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine, University and Regi<strong>on</strong>al Laboratories, Lund, Sweden2 University <str<strong>on</strong>g>of</str<strong>on</strong>g> Campinas ⁄ Hemocentro, Campinas, Brazil3 Bristol Institute for <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Sciences and IBGRL, NHSBT, Bristol, UK4 NIH Clinical Center, Department <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine, Bethesda, MD, USA5 American Red Cross <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Services, Pom<strong>on</strong>a, CA, USA6 New York <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, New York, NY, USA7 LifeShare <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Centers, Shreveport, LA, USA8 Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Laboratory Medicine, Divisi<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> Haematology and <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine, Lund University, Lund, Sweden9 Centre Nati<strong>on</strong>al de Référence pour les Groupes sanguines, Paris, France10 Sanquin Research at CLB, Amsterdam, The Netherlands11 Durban, South Africa12 Osaka Red Cross <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, Osaka, Japan13 Mackay Memorial Hospital and Nati<strong>on</strong>al Taiwan University, Taipei, Taiwan14 <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Bank, Hospital Sirio-Libanes, São Paulo, Brazil15 NBGRL Magen David Adom, Ramat Gan, Israel16 Rh Laboratory, Winnipeg, MB, CanadaReceived: 8 December 2010,accepted 9 December 2010,published <strong>on</strong>line 15 March 2011Key words: blood groups, genetics, terminology.Introducti<strong>on</strong>The <str<strong>on</strong>g>Working</str<strong>on</strong>g> <str<strong>on</strong>g>Party</str<strong>on</strong>g> met in Berlin, Germany during the2010 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> (ISBT)C<strong>on</strong>gress. While no changes were made to the currentclassificati<strong>on</strong> as documented in <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Group Terminology2004 [1] and the 2007 update [2], following a meeting <str<strong>on</strong>g>of</str<strong>on</strong>g>the <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> Scientific Advisory Committee in Cairo,the <str<strong>on</strong>g>Working</str<strong>on</strong>g> <str<strong>on</strong>g>Party</str<strong>on</strong>g> was renamed the <str<strong>on</strong>g>Working</str<strong>on</strong>g> <str<strong>on</strong>g>Party</str<strong>on</strong>g> <strong>on</strong>Red Cell Immunogenetics and <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Group TerminologyCorresp<strong>on</strong>dence: Dr Jill R. Storry, Clinical Immunology & <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>Medicine, University and Regi<strong>on</strong>al Laboratories, Klinikgatan 21, SE-22185Lund, SwedenE-mail: jill.storry@med.lu.seand the following terms <str<strong>on</strong>g>of</str<strong>on</strong>g> reference were drawn up byDr Ge<str<strong>on</strong>g>of</str<strong>on</strong>g>f Daniels:(1) To advise the ISBT <strong>on</strong> matters relating to <strong>red</strong> <strong>cell</strong>immunogenetics.(2) To organize internati<strong>on</strong>al workshops <strong>on</strong> blood groupmolecular genotyping.(3) To maintain and m<strong>on</strong>itor a terminology and geneticclassificati<strong>on</strong> for blood group antigens.(4) To develop, maintain and m<strong>on</strong>itor a terminology forblood group genes and their alleles.A total <str<strong>on</strong>g>of</str<strong>on</strong>g> 14 new blood group antigens were added to 10<str<strong>on</strong>g>of</str<strong>on</strong>g> the 30 current blood group systems (Table 1), and <strong>on</strong>eantigen (FY4) was made obsolete. As described below, anew Collecti<strong>on</strong> c<strong>on</strong>taining the six MNS-related carbohydrateantigens was created. A new antigen, PX2, was addedto Collecti<strong>on</strong> 209, from which the P k antigen was moved to77
78 J. R. Storry et al.AntigennameIncidenceNucleotidechange a Amino acid change ReferenceTable 1 New blood group antigensRH58 CELO High RHCE 697C>G Gln233Glu [7]RH59 CEAG High RHCE 254G>C Ala85Gly [8]LU22 LURC High 223C>T Arg75Cys & dependent <strong>on</strong> Arg77 [9]KEL35 KELP High 780G>T2024G>ALeu260PheArg675GlnDI22 DISK High 1694G>C Gly565Ala [12]DO7 DOMR High 431C>T Ala144Glu [13]432C>ACO4 Co4 High 140A>G Gln47Arg [14,15]GE10 GEPL High 134C>T GPC Pro45Leu; GPD Pro24Leu [16]GE11 GEAT High 56A>T Asp19Val [16]GE12 GETI High 80C>T GPC Thr27Ile; GPD Thr6Ile [16]CROM16 CROZ High 389G>A Arg130His [17]OK2 OKGV High 176G>T Gly59Val [18]OK3 OKVM High 178G>T Val60Met [19]JMH6 JMHQ High 1040G>T Arg347Leu [20][11]a Numbers used for nucleotide and amino acid locati<strong>on</strong> are counted from A <str<strong>on</strong>g>of</str<strong>on</strong>g> the initiating methi<strong>on</strong>inecod<strong>on</strong> and that methi<strong>on</strong>ine residue, respectively.the P blood group system, and was renamed P1PK. Thisbrings the current total <str<strong>on</strong>g>of</str<strong>on</strong>g> recognized blood group antigensto 328, <str<strong>on</strong>g>of</str<strong>on</strong>g> which 284 are c<strong>on</strong>tained within the 30 bloodgroups systems.System 3: P1PKIt has recently been shown that a nucleotide polymorphismupstream <str<strong>on</strong>g>of</str<strong>on</strong>g> the coding regi<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> the A4GALT gene, previouslyshown to encode the 4-a-galactosyltransferase thatsynthesizes the P k antigen [3–5], correlates with P1 antigenexpressi<strong>on</strong> [6]. A novel short transcript <str<strong>on</strong>g>of</str<strong>on</strong>g> A4GALTrevealed a new ex<strong>on</strong> in intr<strong>on</strong> 1, termed ex<strong>on</strong> 2a. Threepolymorphisms were identified in the ex<strong>on</strong>, <str<strong>on</strong>g>of</str<strong>on</strong>g> which <strong>on</strong>enucleotide change correlates with the P 1 ⁄ P 2 phenotypesand for the first time, permits genotyping for P1 antigenexpressi<strong>on</strong> [6]. This finding ties the P1 and P k antigens,l<strong>on</strong>g known to be related serologically and biochemically,to the same gene – A4GALT. Based <strong>on</strong> the molecular evidence(from > 200 d<strong>on</strong>ors) linking the two antigens, it wasproposed to rename the P system to P1PK. This change wasalso based <strong>on</strong> the c<strong>on</strong>fusi<strong>on</strong> that would arise if the P1 andP k antigens but not the P antigen bel<strong>on</strong>ged to a system stillcalled P. The P1 antigen (now P1PK1) retains the ISBTnumber 003001, P k (P1PK2) becomes 003002, while number209002 (formerly P k ) is now obsolete.System 4: RhTwo high-incidence antigens have been added to the Rhsystem. RH58 (CELO) is antithetical to the low-incidenceantigen, RH43 (Crawford), and is not encoded by RHCE*-ceCF (48C, 697G, 733G) and very weakly expressed <strong>on</strong> theDc(e) phenotype encoded by RHCE*ceBP [7]. Two patients,<strong>on</strong>e who was homozygous for RHC E*ceCF the other whohad RHCE*ceCF in trans to a silenced RHCE*cE, had eachmade an anti-Rh17-like antibody that was shown to beanti-CELO. Furthermore, the amino acid changes encodedby RHCE*ceCF (Trp16Cys, Gln233Glu, Leu245Val) give riseto partial c and e antigens, which have permitted the producti<strong>on</strong><str<strong>on</strong>g>of</str<strong>on</strong>g> alloanti-c and alloanti-e, respectively. CELO isalso absent from Rh null ,D)), and DC W ) phenotype RBCs.RH59 (CEAG) is a high-incidence antigen associated withan allele encoding a partial e antigen [8]. It was identifiedfollowing the investigati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> three multiply transfusedpatients whose plasma c<strong>on</strong>tained alloanti-e but whose RBCstyped e+. The absence <str<strong>on</strong>g>of</str<strong>on</strong>g> CEAG is defined by the singlenucleotide change RHCE*ce 254G>C that encodes an aminoacid exchange <str<strong>on</strong>g>of</str<strong>on</strong>g> Ala85Gly.System 5: LutheranA new high-incidence antigen, LU22 (LURC), has beenadded to the Lutheran system. The molecular change resultingin the LU:)22 phenotype was shown to be a nucleotidechange in ex<strong>on</strong> 3, 223C>T that changed Arg75Cys [9]. TheRBCs <str<strong>on</strong>g>of</str<strong>on</strong>g> the proband phenotyped as Lu(a+b+ weak ), andsequencing revealed heterozygosity for this mutati<strong>on</strong> carried<strong>on</strong> a LU*B allele and for an LU*A allele bearing apreviously described Lu a -related polymorphism, 586G>A(Val196Ile) [10]. The Lu a ⁄ Lu b amino acid polymorphismoccurs at residue 77, where Arg77 defines Lu b and His77Ó 2011 The Author(s)Vox Sanguinis Ó 2011 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>, Vox Sanguinis (2011) 101, 77–82
Red <strong>cell</strong> immunogenetics and blood group terminology 79defines Lu a . The proximity <str<strong>on</strong>g>of</str<strong>on</strong>g> Arg75Cys presumably interruptsand weakens the expressi<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> the Lu b antigenencoded by this allele. Furthermore, the occurrence <str<strong>on</strong>g>of</str<strong>on</strong>g> theLURC-negative phenotype <strong>on</strong> a heterozygous LU*A ⁄ LU*Bbackground dem<strong>on</strong>strates the reliance <str<strong>on</strong>g>of</str<strong>on</strong>g> LURC <strong>on</strong> the Lu bantigen residue, Arg77 for expressi<strong>on</strong>.System 6: KellAnother high-incidence antigen was added to the Kell system:KEL35 (KELP), defined by an antibody that was n<strong>on</strong>reactive<strong>on</strong>ly with K 0 RBCs and those <str<strong>on</strong>g>of</str<strong>on</strong>g> the antibody maker[11]. The patient’s RBCs typed KEL:)1,2–3,4,6,11,)12,22,although sequence analysis did not c<strong>on</strong>firm the K:)12phenotype. However, the proband was homozygous fortwo novel mutati<strong>on</strong>s: 780G>T in ex<strong>on</strong> 8, which encodesLeu260Phe, and 2024G>A in ex<strong>on</strong> 18, encoding Arg675Gln. Leu260Phe is located <strong>on</strong> the Kell glycoprotein at apositi<strong>on</strong> distal from the membrane and modelling p<strong>red</strong>ictsa change in the molecular surface <str<strong>on</strong>g>of</str<strong>on</strong>g> Kell. However, aminoacid positi<strong>on</strong> 675 lies close to or in the membrane. Bothmutati<strong>on</strong>s are seemingly unc<strong>on</strong>nected with Arg548 thatdefines K12 and the effect <str<strong>on</strong>g>of</str<strong>on</strong>g> these two novel mutati<strong>on</strong>s <strong>on</strong>K12 expressi<strong>on</strong> remains unresolved.System 8: DuffyFollowing the criteria that an antibody to a blood groupantigen must be extant for that antigen to be recognized,and based <strong>on</strong> <strong>on</strong>ly scant original evidence for its existence,FY4 has been made obsolete.System 10: DiegoA high-incidence antigen antithetical to DI9 (Wu) has beenidentified. The new antigen, DI22 (DISK), was characterizedby an apparently naturally occurring, str<strong>on</strong>gly agglutinatingantibody reactive both at 18 and 37°C and by the indirectantiglobulin test [12]. DISK was shown to be sensitive toa-chymotrypsin treatment, but resistant to other comm<strong>on</strong>lyused proteases. Targeted sequence analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> SLC4A1 ex<strong>on</strong>14 revealed homozygosity in the proband and heterozygosityin a sample from her brother, for the mutati<strong>on</strong> 1694G>Cthat encodes Gly565Ala. The RBCs <str<strong>on</strong>g>of</str<strong>on</strong>g> her brother reactedmore weakly with her antibody, suggesting that the anti-DISK exhibits dosage in DI:9,22 individuals.System 14: DombrockDO7 (DOMR) is defined by an antibody to a high-incidenceantigen produced in a patient, homozygous for two adjacentnucleotide changes 431C>A, 432C>A that encode achange <str<strong>on</strong>g>of</str<strong>on</strong>g> Ala144Glu [13]. These changes are present with aDO*B-WL allele (DO*793G, DO*323G, DO*350C, DO*547T,DO*898G). The DO*B-WL allele is p<strong>red</strong>icted to encode thephenotype Do(a)b+), Hy+, Jo(a+), DOYA+, however, thepatient’s RBCs typed Do(a)b+), Hy+ w , Jo(a+ w ), DOYA+ w .Her RBCs also typed Gy(a)) with a polycl<strong>on</strong>al antibody butwere reactive with several m<strong>on</strong>ocl<strong>on</strong>al antibodies to theDombrock glycoprotein. Further complexity was dem<strong>on</strong>stratedby the plasma antibody that was n<strong>on</strong>reactive or<strong>on</strong>ly very weakly reactive with six Hy) samples, n<strong>on</strong>reactivewith Gy(a)) RBCs but reactive with Jo(a)) and DOYA)RBCs.System 15: COAn antibody compatible with Co(a)b)) RBCs but weaklyincompatible with Co(a)b+) RBCs was identified in theplasma <str<strong>on</strong>g>of</str<strong>on</strong>g> a patient whose RBCs typed Co(a)b)) [14].Molecular analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> AQP1 revealed homozygosity for140A>G, encoding the amino acid change Gln47Arg, inclose proximity to the Co a ⁄ Co b polymorphism at residue45. Samples from a sec<strong>on</strong>d Co(a)b)) proband homozygousfor the same 140A>G mutati<strong>on</strong> were investigated followingdetecti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> an antibody to a high-incidence antigen initiallyidentified as anti-Co3 [15]. Unexpectedly, her RBCswere shown to have functi<strong>on</strong>al aquaporin 1. Expressi<strong>on</strong>studies with AQP1 encoding Arg47 dem<strong>on</strong>strated a loss <str<strong>on</strong>g>of</str<strong>on</strong>g>Co a antigen expressi<strong>on</strong> despite the presence <str<strong>on</strong>g>of</str<strong>on</strong>g> the Co a -specific Ala45, c<strong>on</strong>sistent with the observed phenotype.Arnaud et al. [15] also re-evaluated three antisera classifiedas anti-Co3 in their collecti<strong>on</strong> and re-identified <strong>on</strong>e <str<strong>on</strong>g>of</str<strong>on</strong>g> thethree sera as anti-Co4. PCR–RFLP analysis revealed thatthis sample also was homozygous for AQP1 140G. Thus,this polymorphism defines a new high-incidence antigen,CO4, in the Colt<strong>on</strong> blood group system.System 20: GerbichThree novel high-incidence antigens am<strong>on</strong>g four individualshave been added to the Gerbich blood group system:GE10 (GEPL), GE11 (GEAT), GE12 (GETI) [16]. GE10 wasidentified following the investigati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> an individualwhose RBCs typed GE:2,3,4 but whose plasma c<strong>on</strong>tained anapparent alloanti-Ge3. Immunoblotting analysis showedthe presence <str<strong>on</strong>g>of</str<strong>on</strong>g> both normal GPC ⁄ GPD and two unexpectedbands that were approximately 2 kDa smaller than normal.DNA sequence analysis revealed a single point mutati<strong>on</strong> inGYPC, 134C>T, which encodes an amino acid change Proto Leu at positi<strong>on</strong>s 45 and 24 <str<strong>on</strong>g>of</str<strong>on</strong>g> GPC and GPD, respectively.Anti-Ge11 was n<strong>on</strong>reactive with GE:)2,)3 andGE:)2,)3,)4 RBCs, but dem<strong>on</strong>strated variable reactivitywith GE:)2,3 RBCs. Bands corresp<strong>on</strong>ding to GPC and GPD<str<strong>on</strong>g>of</str<strong>on</strong>g> apparently normal mass were shown by immunoblottingand DNA sequence analysis revealed homozygosity for aÓ 2011 The Author(s)Vox Sanguinis Ó 2011 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>, Vox Sanguinis (2011) 101, 77–82
80 J. R. Storry et al.point mutati<strong>on</strong> 56A>T that encodes an amino acid change<str<strong>on</strong>g>of</str<strong>on</strong>g> Asp19Val in GPC.Anti-Ge12 was identified in the plasma <str<strong>on</strong>g>of</str<strong>on</strong>g> two siblingsand an unrelated individual whose RBCs typed GE:)2,3,4(although typing for Ge3 and Ge4 <strong>on</strong> the siblings’ RBCs wasweaker than c<strong>on</strong>trols). Immunoblotting analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> RBCmembranes from the siblings revealed normal GPC andGPD as well as a broad band similar to that seen inGE:)2,3,4 RBCs, although the RBCs <str<strong>on</strong>g>of</str<strong>on</strong>g> the unrelated patientwere normal. DNA sequence analysis revealed that the siblingsshowed compound heterozygosity for two mutati<strong>on</strong>s:80C>T in ex<strong>on</strong> 2 that encodes Thr27Ile in GPC and Thr6Ilein GPD and a transiti<strong>on</strong> in the invariant ex<strong>on</strong>-adjacentnucleotide <str<strong>on</strong>g>of</str<strong>on</strong>g> intr<strong>on</strong> 1, IVS2 )1g>a. This change most likelycauses ex<strong>on</strong> skipping and results in an aberrant protein,similar to GE:)2,3,4 RBCs in which ex<strong>on</strong> 2 is deleted andexplains the unusual blotting results. The unrelated patientwas homozygous for 80C>T. Thus, Thr27Ile and Thr6Iledefine the GE12 (GETI) antigen.System 21: CromerAn antibody directed at an apparently Cromer-relatedhigh-incidence antigen was shown to detect a new antigenin that system, CROM16 (CROZ) [17]. DNA sequence analysis<str<strong>on</strong>g>of</str<strong>on</strong>g> DAF revealed a missense transiti<strong>on</strong>, 389G>A in ex<strong>on</strong>3 that encodes an amino acid change <str<strong>on</strong>g>of</str<strong>on</strong>g> Arg130His.System 24: OKTwo new high-incidence antigens have been identified in theOK blood group system; OK2 (OKGV) and OK3 (OKVM). TheOK:)2 phenotype described in 2003 was shown to arise froma nucleotide substituti<strong>on</strong> in ex<strong>on</strong> 2 <str<strong>on</strong>g>of</str<strong>on</strong>g> the basigin gene, BSG176G>T, which encodes the amino acid change Gly59Val[18]. This mutati<strong>on</strong> was found <strong>on</strong>ly in the proposita, togetherwith two additi<strong>on</strong>al silent single nucleotide exchanges;195C>T (Asp65) and 234G>C (Ser78). Interestingly, an additi<strong>on</strong>alsilent transiti<strong>on</strong>, 327T>C (Ala108), was identified in<strong>on</strong>e <str<strong>on</strong>g>of</str<strong>on</strong>g> the c<strong>on</strong>trol samples sequenced in this study.Karamatic Crew et al. [19] also determined the molecularbasis <str<strong>on</strong>g>of</str<strong>on</strong>g> the OK:)3 phenotype: the proband’s plasma wasn<strong>on</strong>reactive with three examples <str<strong>on</strong>g>of</str<strong>on</strong>g> Ok(a)) RBCs tested,although her RBCs typed Ok(a+). Sequence analysis <str<strong>on</strong>g>of</str<strong>on</strong>g> BSGrevealed homozygosity for another mutati<strong>on</strong> in ex<strong>on</strong> 2:178G>A, causing a Val60Met change in the OK glycoprotein(basigin, CD147).System 26: JMHA sixth antigen has been added to the increasingly polymorphicJMH blood group system; JMH6 (JMHQ). An antibodywith JMH-related specificity in the plasma <str<strong>on</strong>g>of</str<strong>on</strong>g> fourunrelated native Americans led to the investigati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g>SEMA7A [20]. The molecular basis was identified as atransversi<strong>on</strong>, 1040G>T, which encodes a change <str<strong>on</strong>g>of</str<strong>on</strong>g>Arg347Leu. Recombinant forms <str<strong>on</strong>g>of</str<strong>on</strong>g> SEMA7A were c<strong>on</strong>structedand used in an ELISA, which c<strong>on</strong>firmed reactivity<str<strong>on</strong>g>of</str<strong>on</strong>g> the antibodies with wild-type SEMA7A, while theJMHQ) is<str<strong>on</strong>g>of</str<strong>on</strong>g>orm was n<strong>on</strong>reactive with the plasma samples.Collecti<strong>on</strong> 209: GLOBA new antigen, PX2 has been added to the GLOB Collecti<strong>on</strong>(209). This antigen is a high-incidence antigen defined by aterminal b3-N-acetylgalactosamine (b3GalNAc) <strong>on</strong> paraglobosideand is known to be abundant <strong>on</strong> PP1P k ) (p phenotype)RBCs [21, 22]. Recent data have revealed thatrare individuals lacking the P antigen make antibodiesnot <strong>on</strong>ly against globoside but also against this ‘new’antigen, which makes their plasma incompatible with pphenotype RBCs (J. R. Storry, T. Peyrard, A. H. Hult,Å. Hellberg, M. L. Olss<strong>on</strong>, manuscript in preparati<strong>on</strong>).Collecti<strong>on</strong> 213A new Collecti<strong>on</strong>, number 213 (Table 2), has been createdto recognize the carbohydrate antigens associated with theM and N antigens in MNS (ISBT system 002). Theseantigens have previously been defined biochemically asdifferent substituti<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> the sialic acid-carrying oligosaccharides<strong>on</strong> glycophorin A [23].Allele terminologyTo address the fourth term <str<strong>on</strong>g>of</str<strong>on</strong>g> reference listed at the beginning<str<strong>on</strong>g>of</str<strong>on</strong>g> this article, a subcommittee has drafted guidelinesfor blood group allele terminology that can be found <strong>on</strong> the<str<strong>on</strong>g>Blood</str<strong>on</strong>g> Group Terminology website at http://ibgrl.blood.co.uk/ISBTPages/ISBTHome.htm. Proposed allele terminologyfor 26 <str<strong>on</strong>g>of</str<strong>on</strong>g> the 30 blood group systems can also be foundhere and comments ⁄ feedback are encouraged. The proposedguidelines are intenti<strong>on</strong>ally n<strong>on</strong>inclusive <str<strong>on</strong>g>of</str<strong>on</strong>g> silentsingle nucleotide polymorphisms (SNPs) and otherTable 2 Collecti<strong>on</strong> 213: Antigens that are associated with M and Nantigens and that are the result <str<strong>on</strong>g>of</str<strong>on</strong>g> different substituti<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> the sialicacid-carrying oligosaccharides <strong>on</strong> glycophorin A [23]Number213001 Hu213002 M 1213003 Tm213004 Can213005 Sext213006 SjAntigen nameÓ 2011 The Author(s)Vox Sanguinis Ó 2011 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>, Vox Sanguinis (2011) 101, 77–82
82 J. R. Storry et al.Appendix 1. Members <str<strong>on</strong>g>of</str<strong>on</strong>g> the <str<strong>on</strong>g>Working</str<strong>on</strong>g> <str<strong>on</strong>g>Party</str<strong>on</strong>g>Dr JR Storry (Current chair): Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Clinical Immunologyand <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine, University and Regi<strong>on</strong>alLaboratories, Lund, Sweden (jill.storry@med.lu.se).Dr GL Daniels (Immediate Past Chair): Bristol Institutefor <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Sciences, NHS <str<strong>on</strong>g>Blood</str<strong>on</strong>g> and Transplant,Filt<strong>on</strong>, UK (ge<str<strong>on</strong>g>of</str<strong>on</strong>g>f.daniels@nhsbt.nhs.uk).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr L. Castilho: University <str<strong>on</strong>g>of</str<strong>on</strong>g> Campinas ⁄ Hemocentro,Campinas, Brazil (castilho@unicamp.br).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr WA Flegel: Department <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine,Clinical Center, Nati<strong>on</strong>al Institutes <str<strong>on</strong>g>of</str<strong>on</strong>g> Health, Bethesda, MD,USA (bill.flegel@cc.nih.gov).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr G Garratty: American Red Cross <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Services,Southern California Regi<strong>on</strong>, Pom<strong>on</strong>a, CA, USA (garratty@usa.<strong>red</strong>cross.org).Ms C Lomas-Francis: New York <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, New York,NY, USA (clomas-francis@nybloodcenter.org).Mr JJ Moulds: LifeShare <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Centers, Shreveport, LA,USA (jjmoulds@lifeshare.org).Dr JM Moulds: LifeShare <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Centers, Shreveport, LA,USA (jmmoulds@lifeshare.org).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr ML Olss<strong>on</strong>: Department <str<strong>on</strong>g>of</str<strong>on</strong>g> Laboratory Medicine,Divisi<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> Hematology and <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g> Medicine, LundUniversity, Lund, Sweden (martin_l.olss<strong>on</strong>@med.lu.se).Ms J Poole: IBGRL, NHS <str<strong>on</strong>g>Blood</str<strong>on</strong>g> and Transplant, Bristol,UK (joyce.poole@nhsbt.nhs.uk).Dr ME Reid: New York <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, New York, NY, USA(mreid@nybloodcenter.org).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr Ph Rouger: Centre nati<strong>on</strong>al de Référence pour lesGroupes sanguines, Paris, France (prouger@ints.fr).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr CE van der Schoot: Sanquin Research at CLB,Amsterdam, The Netherlands (e.vanderschoot@sanquin.nl).Pr<str<strong>on</strong>g>of</str<strong>on</strong>g> Dr M Scott: Bristol Institute for <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>Sciences, NHS <str<strong>on</strong>g>Blood</str<strong>on</strong>g> and Transplant, Filt<strong>on</strong>, Bristol, UK(mari<strong>on</strong>.scott@nhsbt.nhs.uk).Mrs E Smart: Durban, South Africa (eapsmart@svnet.co.za).Dr Y Tani: Osaka Red Cross <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, Osaka, Japan(tani@osaka.bc.jrc.or.jp).Dr LC Yu: Mackay Memorial Hospital and Nati<strong>on</strong>alTaiwan University, Taipei, Taiwan (yulc@ntu.edu.tw).Dr S Wendel: <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Bank, Hospital Sirio-Libanes, SãoPaulo, Brazil (snwendel@terra.com.br).Dr CM Westh<str<strong>on</strong>g>of</str<strong>on</strong>g>f: American Red Cross and the University<str<strong>on</strong>g>of</str<strong>on</strong>g> Pennsylvania, Philadelphia, PA, USA., and nowNew York <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Center, New York, NY (cwesth<str<strong>on</strong>g>of</str<strong>on</strong>g>f@nybloodcenter.org).Dr V Yahalom: NBGRL Magen David Adom, Ramat Gan,Israel (ve<strong>red</strong>y@mda.org.il).Dr T Zelinski: Rh Laboratory, Winnipeg, Manitoba,Canada (zelinski@ms.umanitoba.ca).Newly appointed membersDr M de Haas: Sanquin <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Supply Foundati<strong>on</strong>, Amsterdam,The Netherlands (m.dehaas@sanquin.nl).Dr KM Hyland: Australian Red Cross <str<strong>on</strong>g>Blood</str<strong>on</strong>g> Services,Brisbane, Australia (chyland@arcbs.<strong>red</strong>cross.org.au).Dr N Nogues: Banc de Sang i Teixits, Barcel<strong>on</strong>a, Spain(nnogues@bstcat.net).Ó 2011 The Author(s)Vox Sanguinis Ó 2011 <str<strong>on</strong>g>Internati<strong>on</strong>al</str<strong>on</strong>g> <str<strong>on</strong>g>Society</str<strong>on</strong>g> <str<strong>on</strong>g>of</str<strong>on</strong>g> <str<strong>on</strong>g>Blood</str<strong>on</strong>g> <str<strong>on</strong>g>Transfusi<strong>on</strong></str<strong>on</strong>g>, Vox Sanguinis (2011) 101, 77–82