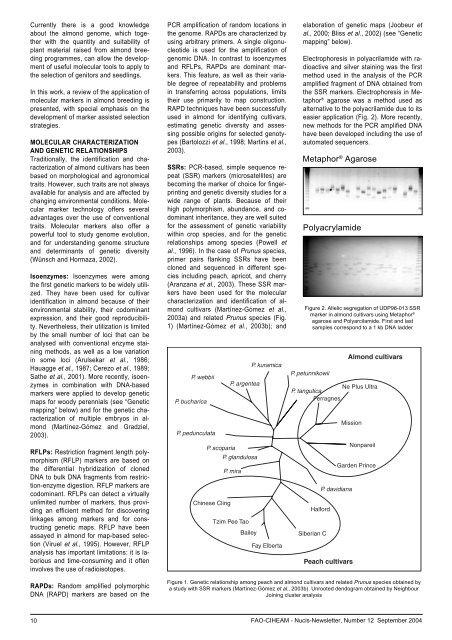
Currently there is a good knowledgeabout the almond genome, which togetherwith the quantity and suitability ofplant material raised from almond breedingprogrammes, can allow the developmentof useful molecular tools to apply tothe selection of genitors and seedlings.In this work, a review of the application ofmolecular markers in almond breeding ispresented, with special emphasis on thedevelopment of marker assisted selectionstrategies.MOLECULAR CHARACTERIZATIONAND GENETIC RELATIONSHIPSTraditionally, the identification and characterizationof almond cultivars has beenbased on morphological and agronomicaltraits. However, such traits are not alwaysavailable for analysis and are affected bychanging environmental conditions. Molecularmarker technology offers severaladvantages over the use of conventionaltraits. Molecular markers also offer apowerful tool to study genome evolution,and for understanding genome structureand determinants of genetic diversity(Wünsch and Hormaza, 2002).Isoenzymes: Isoenzymes were amongthe first genetic markers to be widely utilized.They have been used for cultivaridentification in almond because of theirenvironmental stability, their codominantexpression, and their good reproducibility.Nevertheless, their utilization is limitedby the small <strong>number</strong> of loci that can beanalysed with conventional enzyme stainingmethods, as well as a low variationin some loci (Arulsekar et al., 1986;Hauagge et al., 1987; Cerezo et al., 1989;Sathe et al., 2001). More recently, isoenzymesin combination with DNA-basedmarkers were applied to develop geneticmaps for woody perennials (see “Geneticmapping” below) and for the genetic characterizationof multiple embryos in almond(Martínez-Gómez and Gradziel,2003).RFLPs: Restriction fragment length polymorphism(RFLP) markers are based onthe differential hybridization of clonedDNA to bulk DNA fragments from restriction-enzymedigestion. RFLP markers arecodominant. RFLPs can detect a virtuallyunlimited <strong>number</strong> of markers, thus providingan efficient method for discoveringlinkages among markers and for constructinggenetic maps. RFLP have beenassayed in almond for map-based selection(Viruel et al., 1995). However, RFLPanalysis has important limitations: it is laboriousand time-consuming and it ofteninvolves the use of radioisotopes.PCR amplification of random locations inthe genome. RAPDs are characterized byusing arbitrary primers. A single oligonucleotideis used for the amplification ofgenomic DNA. In contrast to isoenzymesand RFLPs, RAPDs are dominant markers.This feature, as well as their variabledegree of repeatability and problemsin transferring across populations, limitstheir use primarily to map construction.RAPD techniques have been success<strong>full</strong>yused in almond for identifying cultivars,estimating genetic diversity and assessingpossible origins for selected genotypes(Bartolozzi et al., 1998; Martins et al.,2003).SSRs: PCR-based, simple sequence repeat(SSR) markers (microsatellites) arebecoming the marker of choice for fingerprintingand genetic diversity studies for awide range of plants. Because of theirhigh polymorphism, abundance, and codominantinheritance, they are well suitedfor the assessment of genetic variabilitywithin crop species, and for the geneticrelationships among species (Powell etal., 1996). In the case of Prunus species,primer pairs flanking SSRs have beencloned and sequenced in different speciesincluding peach, apricot, and cherry(Aranzana et al., 2003). These SSR markershave been used for the molecularcharacterization and identification of almondcultivars (Martínez-Gómez et al.,2003a) and related Prunus species (Fig.1) (Martínez-Gómez et al., 2003b); andelaboration of genetic maps (Joobeur etal., 2000; Bliss et al., 2002) (see “Geneticmapping” below).Electrophoresis in polyacrilamide with radioactiveand silver staining was the firstmethod used in the analysis of the PCRamplified fragment of DNA obtained fromthe SSR markers. Electrophoresis in Metaphor® agarose was a method used asalternative to the polyacrilamide due to itseasier application (Fig. 2). More recently,new methods for the PCR amplified DNAhave been developed including the use ofautomated sequencers.Metaphor ® AgarosePolyacrylamideFigure 2. Allelic segregation of UDP96-013 SSRmarker in almond cultivars using Metaphor ®agarose and Polyarcilamide. First and lastsamples correspond to a 1 kb DNA ladderRAPDs: Random amplified polymorphicDNA (RAPD) markers are based on theFigure 1. Genetic relationship among peach and almond cultivars and related Prunus species obtained bya study with SSR markers (Martínez-Gómez et al., 2003b). Unrooted dendogram obtained by NeighbourJoining cluster analysis10 FAO-CIHEAM - Nucis-Newsletter, Number 12 <strong>September</strong> 2004
GENETIC MAPPINGSeveral intraspecific and interspecific almondmaps have been developed usingdifferent types of molecular markers. Theuse of PCR-based markers made it possibleto map and tag a wide range of traits(Arús et al., 1994; 1999). The analysis ofcosegregation among markers eases linkageanalysis between markers and majoror quantitative loci controlling horticulturallyimportant traits.Different research groups have releasedlinkage maps using morphological traits,isoenzymes, RFLPs and RAPDs in almond(Viruel et al., 1995; Joobeur et al.,1998; 2000; Jáuregui et al., 2001; Bliss etal., 2002). In addition, SSRs have beenused for mapping in almond (Joobeur etal., 2000; Bliss et al., 2002; Aranzana etal., 2003). The similar arrangement ofmarkers observed in different Prunusmaps suggests a high level of syntenywithin the genus (Aranzana et al., 2003).This homology among Prunus speciespartly explains the low level of breedingbarriers to interspecific gene introgressionand supports the opportunity for successfulgene transfer between closely relatedspecies (Gradziel et al., 2001).MARKER-ASSISTED SELECTIONSelection by molecular markers is particularlyuseful in fruit, nut, and other treecrops with a long juvenile period, andwhen the expression of the gene is recessiveor the assessment of the character isdifficult. Marker-assisted selection (MAS)is emerging as a very promising strategyfor increasing selection gains. If sufficientmapping information is available, MAScan highly shorten the <strong>number</strong> of generationsrequired to “eliminate” the undesiredgenes of the donor in backcrossing programmes(Arús and Moreno-González,1993). Marker loci linked to major genescan be used for selection, which is moreefficient than direct selection for the targetgene (Arús and Moreno-González,1993).The principal approach for the analysis ofmarker-trait association in almond is theuse of mapping populations segregatingfor the characters of interest. The differentlinkage maps developed in almondinclude markers associated with severaltraits of horticultural value. MappingTable 1. Markers associated to main agronomic traits in almond.quantitative characters by identifyingquantitative trait loci (QTL) is also becomingan important tool in tree breeding.QTLs are generally recognized by comparingthe degree of covariation for polymorphicmolecular markers with phenotypictrait measurements. Important charactersand QTLs that are presently beingmapped in almond include bloom time,self-incompatibility, shell hardness, orkernel taste (Asins et al., 1994; Viruel etal., 1995; Ballester et al., 1998, 2001;Jobeur et al., 1998; 2002; Bliss et al.,2002) (Table 1). The high degree of genomesynteny observed among Prunusspecies should also facilitate the successfultransfer of sets of markers and codingsequence among species (Aranzanaet al., 2003) (Table 1).Bulk segregant analysis (BSA), wheretwo pooled DNA samples are formed fromplant sources that have similar geneticbackgrounds but differ in one particulartrait, is another promising approach forthe analysis of molecular marker-horticulturaltrait association. A strategy combiningdifferent markers with bulk segregantanalysis was used to identify markerslinked to loci of specific characters inpeach x almond crosses (Warburton etal., 1996), and three RAPD markers associatedwith a gene conferring delayedbloom in almond (Ballester et al., 2001)(Table 1).A very promising application of molecularmarker assisted selection is the manipulationof self-compatibility in almond. Thisspecies is predominantly self-incompatible.Self-incompatibility is of the gametophytictype and acts to prevent self-fertilization.This character is controlled by asingle locus with multiple codominantalleles (Dicenta and García, 1993), and isexpressed within the styles of flowers asS-RNases glycoproteins (Bošković et al.,1997; 2003) that are responsible for thesubsequent inactivation of self-pollentube growth. Almond self-incompatibilityalleles (S-alleles) were initially identifiedin the field through controlled crosseswith a series of known S-genotypes (Kesterand Gradziel, 1996). More recently,molecular methods have been developedin two areas: identification of stylar S-RNases by electrophoresis in verticalpolyacrilamide gels (Bošković et al.,Trait Symbol Marker ReferenceSelf-incompatibility SI RFLP Joobeur et al. 1998Self-compatibility Sf RFLP Arús et al. 1999Kernel taste Sw RFLP Bliss et al. 2002Shell hardiness D RFLP Arús et al. 1999Late blooming Lb RAPD Ballester et al. 20011997, 2003), and the amplification of specificS-alleles using appropriately designedprimers for PCR and electrophoresisin horizontal agarose gels (Tamura et al.,2000; Channuntapipat et al., 2003). Thistechnique is being routinely used for theidentification of cross-incompatibility groupingsfor current almond cultivars and forefficiently breeding self-compatibility intonew cultivars (Gradziel et al., 2001b; Ortegaand Dicenta, 2003).ACKNOWLEDGMENTSThe authors acknowledge the support ofSpanish Ministry of Education and Science.BIBLIOGRAPHYAranzana M.J., Pineda A., Cosson P.,Dirlewanger E., Ascasibar J., CiprianiG., Ryder C.D., Testolin R., Abbott A.,King G.J., Lezzoni A.F. and P. Arús.2003. A set of simple-sequence (SSR)markers covering the Prunus genome.Theor. Appl. Genet. 106: 819-825.Arulsekar S., Parfitt D.E. and D.E. Kester.1986. Comparison of isozyme variabilityin peach and almond cultivars. J.Hered. 77: 272-274.Arús P. and J. Moreno-González. 1993.Marker-assisted selection. In: Hayward,M.D., Bosemark, N.O., and Romagosa,I. (eds.). Plant Breeding. Principles andProspects. London, United Kingdom:Chapman & Hall. p. 314-331.Arús P., Messeguer R., Viruel M.A., TobuttK., Dirlewanger E., Santi F., QuartaR. and E. Ritter. 1994. The EuropeanPrunus mapping project. Euphytica 77:97-100.Arús P., Ballester J., Jauregui B., JoobeurT., Truco M.J. and M.C. de Vicente.1999. The European Prunus mappingproject: update of marker developmentin almond. Acta Hort. 484: 331-336.Asins M.J., Mestre, P., García, J.E., Dicenta,F., and E.A. Carbonell. 1994.Genotype ∞ environment interaction inQTL analysis of an intervarietal almondcross by means of genetic markers.Theor. Appl. Genet. 89: 358-364.Ballester J., Bošković R., Batlle I., ArúsP., Vargas F. and M.C. de Vicente.1998. Location of the self-incompatibilitygene on the almond linkage map.Plant Breed. 117: 69-72.Ballester J., Socias i Company R., ArúsP. and M.C. de Vicente. 2001. Geneticmapping of a major gene delayingblooming time in almond. Plant Breed.120: 268-270.Bartolozzi F., Warburton M.L., ArulsekarS. and T.M. Gradziel. 1998. Geneticcharacterization and relatedness amongCalifornia almond cultivars and breedinglines detected by randomly amplifiedpolymorphic DNA (RAPD) analysis.J. Am. Soc. Hort. Sci. 123: 381-387.FAO-CIHEAM - Nucis-Newsletter, Number 12 <strong>September</strong> 200411