Electrophoretic characterization of Amaranthus L. seed proteins and ...
Electrophoretic characterization of Amaranthus L. seed proteins and ...
Electrophoretic characterization of Amaranthus L. seed proteins and ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
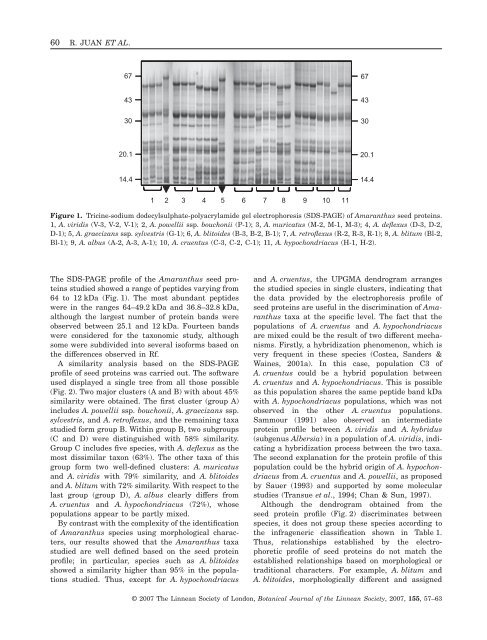
60 R. JUAN ET AL.67674330433020.120.114.414.41 2 3 4 5 6 7 8 9 10 11Figure 1. Tricine-sodium dodecylsulphate-polyacrylamide gel electrophoresis (SDS-PAGE) <strong>of</strong> <strong>Amaranthus</strong> <strong>seed</strong> <strong>proteins</strong>.1, A. viridis (V-3, V-2, V-1); 2, A. powellii ssp. bouchonii (P-1); 3, A. muricatus (M-2, M-1, M-3); 4, A. deflexus (D-3, D-2,D-1); 5, A. graecizans ssp. sylvestris (G-1); 6, A. blitoides (B-3, B-2, B-1); 7, A. retr<strong>of</strong>lexus (R-2, R-3, R-1); 8, A. blitum (Bl-2,Bl-1); 9, A. albus (A-2, A-3, A-1); 10, A. cruentus (C-3, C-2, C-1); 11, A. hypochondriacus (H-1, H-2).The SDS-PAGE pr<strong>of</strong>ile <strong>of</strong> the <strong>Amaranthus</strong> <strong>seed</strong> <strong>proteins</strong>studied showed a range <strong>of</strong> peptides varying from64 to 12 kDa (Fig. 1). The most abundant peptideswere in the ranges 64–49.2 kDa <strong>and</strong> 36.8–32.8 kDa,although the largest number <strong>of</strong> protein b<strong>and</strong>s wereobserved between 25.1 <strong>and</strong> 12 kDa. Fourteen b<strong>and</strong>swere considered for the taxonomic study, althoughsome were subdivided into several is<strong>of</strong>orms based onthe differences observed in Rf.A similarity analysis based on the SDS-PAGEpr<strong>of</strong>ile <strong>of</strong> <strong>seed</strong> <strong>proteins</strong> was carried out. The s<strong>of</strong>twareused displayed a single tree from all those possible(Fig. 2). Two major clusters (A <strong>and</strong> B) with about 45%similarity were obtained. The first cluster (group A)includes A. powellii ssp. bouchonii, A. graecizans ssp.sylvestris, <strong>and</strong> A. retr<strong>of</strong>lexus, <strong>and</strong> the remaining taxastudied form group B. Within group B, two subgroups(C <strong>and</strong> D) were distinguished with 58% similarity.Group C includes five species, with A. deflexus as themost dissimilar taxon (63%). The other taxa <strong>of</strong> thisgroup form two well-defined clusters: A. muricatus<strong>and</strong> A. viridis with 79% similarity, <strong>and</strong> A. blitoides<strong>and</strong> A. blitum with 72% similarity. With respect to thelast group (group D), A. albus clearly differs fromA. cruentus <strong>and</strong> A. hypochondriacus (72%), whosepopulations appear to be partly mixed.By contrast with the complexity <strong>of</strong> the identification<strong>of</strong> <strong>Amaranthus</strong> species using morphological characters,our results showed that the <strong>Amaranthus</strong> taxastudied are well defined based on the <strong>seed</strong> proteinpr<strong>of</strong>ile; in particular, species such as A. blitoidesshowed a similarity higher than 95% in the populationsstudied. Thus, except for A. hypochondriacus<strong>and</strong> A. cruentus, the UPGMA dendrogram arrangesthe studied species in single clusters, indicating thatthe data provided by the electrophoresis pr<strong>of</strong>ile <strong>of</strong><strong>seed</strong> <strong>proteins</strong> are useful in the discrimination <strong>of</strong> <strong>Amaranthus</strong>taxa at the specific level. The fact that thepopulations <strong>of</strong> A. cruentus <strong>and</strong> A. hypochondriacusare mixed could be the result <strong>of</strong> two different mechanisms.Firstly, a hybridization phenomenon, which isvery frequent in these species (Costea, S<strong>and</strong>ers &Waines, 2001a). In this case, population C3 <strong>of</strong>A. cruentus could be a hybrid population betweenA. cruentus <strong>and</strong> A. hypochondriacus. This is possibleas this population shares the same peptide b<strong>and</strong> kDawith A. hypochondriacus populations, which was notobserved in the other A. cruentus populations.Sammour (1991) also observed an intermediateprotein pr<strong>of</strong>ile between A. viridis <strong>and</strong> A. hybridus(subgenus Albersia) in a population <strong>of</strong> A. viridis, indicatinga hybridization process between the two taxa.The second explanation for the protein pr<strong>of</strong>ile <strong>of</strong> thispopulation could be the hybrid origin <strong>of</strong> A. hypochondriacusfrom A. cruentus <strong>and</strong> A. powellii, as proposedby Sauer (1993) <strong>and</strong> supported by some molecularstudies (Transue et al., 1994; Chan & Sun, 1997).Although the dendrogram obtained from the<strong>seed</strong> protein pr<strong>of</strong>ile (Fig. 2) discriminates betweenspecies, it does not group these species according tothe infrageneric classification shown in Table 1.Thus, relationships established by the electrophoreticpr<strong>of</strong>ile <strong>of</strong> <strong>seed</strong> <strong>proteins</strong> do not match theestablished relationships based on morphological ortraditional characters. For example, A. blitum <strong>and</strong>A. blitoides, morphologically different <strong>and</strong> assigned© 2007 The Linnean Society <strong>of</strong> London, Botanical Journal <strong>of</strong> the Linnean Society, 2007, 155, 57–63