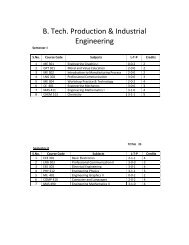
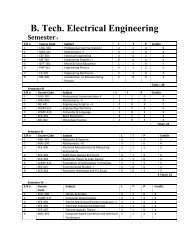
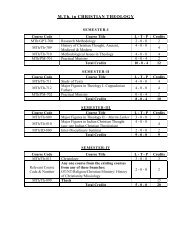
B.Tech. Biotechnology (Molecular & Cellular ... - Shiats.edu.in
B.Tech. Biotechnology (Molecular & Cellular ... - Shiats.edu.in
B.Tech. Biotechnology (Molecular & Cellular ... - Shiats.edu.in
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Introduction: Def<strong>in</strong>ition of Bio<strong>Molecular</strong> model<strong>in</strong>g, Prote<strong>in</strong> Structure – Primary,Secondary and tertiary, Structure of RNA, Nucleotide, am<strong>in</strong>o acids.Unit 2Biological Databases: Types of databases and their <strong>in</strong>troduction. Primary,Secondary, Structure, Ligand Databases.Unit 3Prote<strong>in</strong> Structure determ<strong>in</strong><strong>in</strong>g methods: X-Ray, NMR, RNA Structure Prediction.Unit 4In silico methods of prote<strong>in</strong> structure prediction: Thread<strong>in</strong>g, Ab-<strong>in</strong>itio, homologyModel<strong>in</strong>g.Unit 5Evaluation of modeled prote<strong>in</strong> structure: Ramachandran plot, What Check,ProCheck, Verify 3-D.Unit 6Model<strong>in</strong>g Prote<strong>in</strong> <strong>in</strong>teraction: Prote<strong>in</strong> – Prote<strong>in</strong> Interaction, Prote<strong>in</strong> – LigandInteraction, Software.Unit 7Application of Bio<strong>Molecular</strong> model<strong>in</strong>g.Practicals:1. Introduction to the structure database PDB.2. Visualization of the prote<strong>in</strong> structure us<strong>in</strong>g VMD.3. Secondary structure prediction us<strong>in</strong>g GOR algorithm.4. Tertiary structure prediction us<strong>in</strong>g SWISS-MODEL, ModWeb and Geno3D.5. Us<strong>in</strong>g Modeller for homology model<strong>in</strong>g.6. Prote<strong>in</strong> Structure validation us<strong>in</strong>g SAVS server.7. Prote<strong>in</strong> active site prediction us<strong>in</strong>g CastP and Pocket F<strong>in</strong>der.8. Chemical file format conversion us<strong>in</strong>g Open Babel.9. Automated dock<strong>in</strong>g us<strong>in</strong>g PATCH DOCK webserver.10. Structural alignment us<strong>in</strong>g DaliLite and SSAP.JSBB-589 SEMINAR 1(0-0-1)JSBB-500 TRAINING II EVALUATION 1(0-0-1)