Split-read Analysis Paired-end Mapping Analysis
Split-read Analysis Paired-end Mapping Analysis
Split-read Analysis Paired-end Mapping Analysis
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
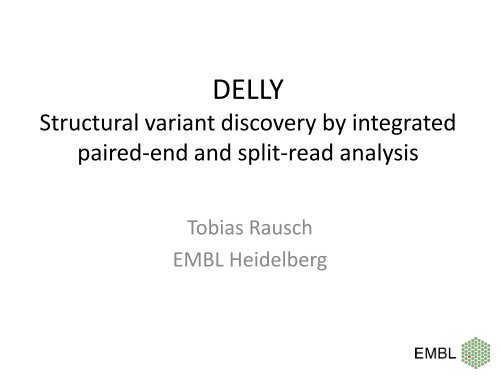
DELLY<br />
Structural variant discovery by integrated<br />
paired-<strong>end</strong> and split-<strong>read</strong> analysis<br />
Tobias Rausch<br />
EMBL Heidelberg
Structural Variants (SVs) discovered by DELLY
SV Detection Methods<br />
(1) <strong>Paired</strong>-<strong>end</strong> mapping (2) <strong>Split</strong>-<strong>read</strong> alignment
SV Detection Methods<br />
(3) Read-depth (4) Assembly
Deletion<br />
Short insertion<br />
(< Insert Size)<br />
Large insertion<br />
(> Insert Size)<br />
Inversion<br />
Tandem duplication<br />
Translocation<br />
Gain/Loss (CNVs)<br />
SV Detection Methods<br />
<strong>Paired</strong>-<strong>end</strong> mapping Read-depth <strong>Split</strong>-<strong>read</strong> Local assembly<br />
Region / Breakpoint Region Region Breakpoint Breakpoint<br />
Delly: Integrated SV Detection
<strong>Paired</strong>-<strong>end</strong> <strong>Mapping</strong><br />
<strong>Analysis</strong><br />
<strong>Split</strong>-<strong>read</strong><br />
<strong>Analysis</strong><br />
Library1.bam<br />
Median: 250bp<br />
MAD: 30bp<br />
Layout:<br />
Delly Components<br />
Library2.bam<br />
Median: 4500bp<br />
MAD: 400bp<br />
Layout:<br />
Deletions at breakpoint level<br />
LibraryN.bam<br />
Median: X bp<br />
MAD: Y bp<br />
Layout:
SV <strong>Paired</strong>-<strong>end</strong> Signatures
<strong>Paired</strong>-<strong>end</strong> Clustering<br />
• <strong>Paired</strong>-<strong>end</strong> Clustering<br />
� Nodes equal discordant paired-<strong>end</strong>s<br />
� Edges connect paired-<strong>end</strong>s that support the same SV<br />
� Edge weights indicate level of agreement<br />
� Graph-based SV Clustering Algorithm<br />
� Connected components<br />
� Heuristic maximal clique identification
<strong>Split</strong>-<strong>read</strong><br />
Search
Low resolution<br />
SV predictions<br />
DELLY: Recent Extensions<br />
Library1.bam Library2.bam LibraryN.bam<br />
<strong>Paired</strong>-<strong>end</strong> <strong>Mapping</strong> <strong>Analysis</strong><br />
<strong>Split</strong>-<strong>read</strong> <strong>Analysis</strong><br />
SVs at breakpoint<br />
resolution<br />
<strong>Paired</strong>-<strong>end</strong><br />
predicted SV calls
Results<br />
(1) Simulations (2) 1000 Genomes Project (3) PCR<br />
Pilot Phase:<br />
8384 assembled deletions<br />
-Delly recovered 76%<br />
Phase I:<br />
-Delly predictions are part of<br />
Phase I deletion callset<br />
ECCB 2012 Paper<br />
DELLY: Structural variant discovery by integrated paired-<strong>end</strong> and split-<strong>read</strong> analysis.<br />
Tobias Rausch, Thomas Zichner, Andreas Schlattl, Adrian Stütz, Vladimir Benes and Jan Korbel.
Application: Filtering of cancer-specific SVs using Germline/1kGP data<br />
• Filtering based-upon call properties<br />
• <strong>Paired</strong>-<strong>end</strong> support<br />
• <strong>Mapping</strong> quality<br />
• <strong>Split</strong>-<strong>read</strong> support
• Filtering against<br />
• 1000 Genomes Project<br />
• Blood/control samples<br />
Annotation/Filtering of SV Calls
Application: Medulloblastoma SV <strong>Analysis</strong><br />
Harboring Complex DNA alterations<br />
Log 2 <strong>read</strong>-depth ratio<br />
Complex alterations
Complex DNA alterations forming<br />
double-minute chromosome
Validation of double-minute and co-<br />
localization of distant segments<br />
Inter-chromosomal connections<br />
validated by PCR<br />
Co-localization of distal<br />
segments on chr3 by FISH
Progressive mutation<br />
acquisition model<br />
Chromothripsis model<br />
Stephens et al., Cell, 2011; Rausch et al., Cell, 2012
EMBL Korbel group<br />
Thomas Zichner<br />
Andreas Schlattl<br />
Adrian Stütz<br />
Jan Korbel<br />
DKFZ<br />
David Jones<br />
Marc Zapatka<br />
Natalie Jäger<br />
Peter Lichter<br />
Stefan Pfister<br />
1000 Genomes Project<br />
ICGC Project Members<br />
Acknowledgements<br />
Thank You!<br />
EMBL Genecore<br />
Markus Fritz<br />
Vladimir Benes<br />
EMBL Huber group<br />
Julian Gehring<br />
Paul Theodor Pyl<br />
Wolfgang Huber<br />
EMBL Steinmetz group<br />
Jonathan Landry<br />
DELLY website: www.embl.de/~rausch/delly.html<br />
rausch@embl.de