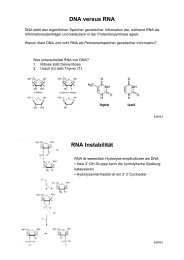
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
ZMBH J.Bericht 2000 - Zentrum für Molekulare Biologie der ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Abbildung auf dem Umschlag:<br />
Das Titelbild zeigt eine symbolische Darstellung des<br />
“funktionalen Genomik-Ansatzes” am Beispiel von<br />
Mycoplasma pneumoniae. Kolorierte elektronenmikroskopische<br />
Aufnahmen von Mycoplasma pneumoniae<br />
bilden den Hintergrund. Die Genkarte im <strong>Zentrum</strong><br />
mit farbkodierten Genfunktionen und <strong>der</strong>en zirkuläre<br />
Darstellung in <strong>der</strong> rechten oberen Ecke bilden<br />
den Ausgangspunkt <strong>für</strong> die Transkriptionsanalyse mittels<br />
Mikro-Array-Technik (mitte rechts) und die Proteomik<br />
(unten rechts). Ein Ausschnitt aus einem zweidimensionalen<br />
Gel, integriert in ein Massenspektrum,<br />
stellen den Ansatz <strong>der</strong> Proteom-Analyse dar.<br />
Cover illustration:<br />
The cover depicts an illustrative representation of the<br />
functional genomics approach on Mycoplasma pneumoniae.<br />
The background is showing electromicrographs<br />
of Mycoplasma pneumoniae cells. The genemap<br />
in the center with a colour coding for gene functions<br />
and in the right corner a segment from a circular<br />
gene-map are both serving as basis for transcription<br />
analysis by micro-array (center right) and for proteomics<br />
(bottom right). The proteomics approach is<br />
represented by a two-dimensional gel integrated in a<br />
mass spectrum, showing partial sequence of a tryptic<br />
peptide.<br />
J. Regula, C.-U. Zimmermann, Y. Cully<br />
and R. Herrmann<br />
<strong>Zentrum</strong> <strong>für</strong> <strong>Molekulare</strong> <strong>Biologie</strong> • Universität Heidelberg (<strong>ZMBH</strong>)<br />
<strong>ZMBH</strong><br />
Im Neuenheimer Feld 282<br />
Postfach 10 62 49<br />
D-69120 Heidelberg<br />
Telefon (0 62 21) 54 68 00<br />
Telefax (00 49 - 62 21) 54 55 07<br />
Internet http://www.zmbh.uni-heidelberg.de<br />
Report <strong>2000</strong>
<strong>ZMBH</strong> Report <strong>2000</strong><br />
Verantwortlich: Prof. Dr. Dr. h.c. Konrad Beyreuther<br />
Redaktion: Prof. Dr. Dr. h.c. Konrad Beyreuther<br />
Dr. H. P. Blaschkowski<br />
Gerlind Güth-Köhler<br />
Gesamtherstellung: Yves Cully<br />
2<br />
Inhaltsverzeichnis - Table of Contents<br />
Forschungsgruppenleiter, Direktorium 6<br />
Wissenschaftlicher Beirat - Scientific Advisory Board 7<br />
Vorbemerkungen des Direktors - Introductory Remarks of the Director 8<br />
Research Reports 23<br />
Konrad Beyreuther Molecular Neurobiology of Alzheimer's Disease 25<br />
Gerd Multhaup 1 Ligand-Dependent Functions of the Amyloid Precursor Protein 40<br />
(APP) and Ligand-Associated Conformational Changes<br />
Hermann Bujard I. Expanding the Applicability of the Tet Regulatory Systems 48<br />
II. The Merozoite Surface Protein 1 of the Human Malaria<br />
Parasite Plasmodium falciparum<br />
Christine Clayton Molecular Cell Biology of Trypanosomes 54<br />
Bernhard Dobberstein Protein Targeting and Intracellular Sorting 61<br />
Dirk Görlich Nucleocytoplasmic Transport 67<br />
Richard Herrmann Molecular Biology of the Bacterium Mycoplasma pneumoniae 74<br />
Ralf-Peter Jansen Asymmetric Cell Division and RNA Transport in Yeast 80<br />
Stefan Jentsch Ubiquitin-Dependent Proteolysis 84<br />
Jörg Höhfeld 1 Function and Regulation of the Mammalian Chaperone Hsc70 88<br />
Klaus-Armin Nave Myelin Genetics and Developmental Neurobiology 91<br />
Renato Paro Chromatin-Controlled Epigenetic Regulation of Transcription 97<br />
3
Frank Sauer Mechanisms of Transcriptional Regulation 104<br />
Hans Ulrich Schairer I. Stigmatella aurantiaca, a Prokaryotic Organism for Studying 109<br />
Intercellular Signalling and Morphogenesis<br />
II. Molecular Biology of the Infection Process by the Entomo-<br />
pathogenic Fungus Beauveria bassiana<br />
Heinz Schaller Regulation of Hepatitis B Virus Replication 116<br />
Percy Knolle 1 Regulation of the Immune Response in the Liver 125<br />
Blanche Schwappach Quality Control of Ion Channels and ABC Proteins 129<br />
Dominique Soldati-Favre Cell and Molecular Biology of the Obligate Intracellular Parasite 131<br />
Toxoplasma gondii<br />
_____________<br />
1 Project Group<br />
Central Facilities 139<br />
<strong>ZMBH</strong>-Lehrprogramm im Grund- und Hauptstudium 148<br />
Studienprogramm <strong>für</strong> Graduierte des <strong>ZMBH</strong> 152<br />
Anhang - Appendix 157<br />
<strong>ZMBH</strong>-Colloquia, Seminars and Cell Biology Lectures 1998/99 – Invited Speakers 158<br />
Administrative and Technical Staff 164<br />
<strong>ZMBH</strong>-Budget 1999 165<br />
4
Forschungsgruppenleiter<br />
Konrad Beyreuther, Prof. Dr. Dr. h.c.<br />
Tel.: 06221-546845, Fax: 06221-545891<br />
Hermann Bujard, Prof. Dr. Dr. h.c<br />
Tel.: 06221-548215, Fax: 06221-545892<br />
Christine Clayton, Prof. Dr.<br />
Tel.: 06221-546876, Fax: 06221-545894<br />
Bernhard Dobberstein, Prof. Dr.<br />
Tel.: 06221-546825, Fax: 06221-545892<br />
Dirk Görlich, Dr.<br />
Tel.: 06221-545884, Fax: 06221-545892<br />
Richard Herrmann, Prof. Dr.<br />
Tel.: 06221-546827, Fax: 06221-545893<br />
Ralf-Peter Jansen, Dr.<br />
Tel.: 06221-546869, Fax: 06221-545894<br />
Stefan Jentsch, Prof. Dr. 1)<br />
Klaus-Armin Nave, Prof. Dr. 2)<br />
Renato Paro, Prof. Dr.<br />
Tel.: 06221-546878, Fax: 06221-545891<br />
Frank Sauer, Dr.<br />
Tel.: 06221-546858, Fax: 06221-545894<br />
Hans Ulrich Schairer, Prof. Dr.<br />
Tel.: 06221-546880, Fax: 06221-545893<br />
Heinz Schaller, Prof. Dr.<br />
Tel.: 06221-546885, Fax: 06221-545893<br />
Dominique Soldati-Favre, Dr.<br />
Tel.: 06221-546870, Fax: 545892<br />
Projektgruppenleiter<br />
Jörg Höhfeld, PD Dr. 3)<br />
Percy Knolle, PD Dr.<br />
Tel.: 06221-546815, Fax: 06221-545893<br />
Gerd Multhaup, PD Dr.<br />
Tel.: 06221-546849, Fax: 06221-545891<br />
Direktorium<br />
Prof. Dr.Dr. h.c. Konrad Beyreuther (Direktor)<br />
Prof. Dr. Klaus-Armin Nave (1. Stellvertreter)<br />
Prof. Dr. Bernhard Dobberstein (2. Stellvertreter)<br />
Sekretariat: Dr. Hans Peter Blaschkowski<br />
Gerlind Güth-Köhler<br />
Tel.: 06221-546850, Fax: 06221-545507<br />
1) Neue Adresse: Max-Planck-Institut <strong>für</strong> Biochemie,<br />
Am Klopferspitz 18a, 82152 Martinsried,<br />
Tel.: 089-8578-3000/9, Fax: 089-8578-3011/3022<br />
2) Neue Adresse: Max-Planck-Institut <strong>für</strong> Experimentelle<br />
Medizin, Abt. Neurogenetik, Herrmann-Rein-<br />
Str. 3, 37075 Göttingen, Tel.: 0551-3899757,<br />
Fax: 0551-3899758<br />
3) Neue Adresse: Max-Planck-Institut <strong>für</strong> Biochemie,<br />
Abt. <strong>Molekulare</strong> Zellbiologie, Am Klopferspitz<br />
18a, 82152 Martinsried, Tel. : 089-8578-3027,<br />
Fax: 089-8578-3022<br />
Wissenschaftlicher Beirat – Scientific Advisory Board<br />
Prof. Dr. Volkmar Braun<br />
Institut <strong>für</strong> <strong>Biologie</strong> II<br />
Universität Tübingen<br />
Prof. Dr. Walter Doerfler<br />
Institut <strong>für</strong> Genetik<br />
Universität Köln<br />
Prof. Dr. Kurt von Figura<br />
Abteilung Biochemie II<br />
Universität Göttingen<br />
Prof. Dr. Ari Helenius<br />
Lab. for Biochemistry<br />
Swiss Fe<strong>der</strong>al Institute<br />
of Technology Zürich<br />
Prof. Dr. Peter Herrlich<br />
Institut <strong>für</strong> Genetik und Toxikologie<br />
von Spaltstoffen<br />
Forschungszentrum Karlsruhe<br />
Prof. Dr. Harvey Lodish<br />
Whitehead Institute<br />
for Biomedical Research<br />
Cambridge, MA<br />
Prof. Dr. Richard Losick<br />
Dept. of Cellular & Developmental Biology<br />
Harvard University<br />
Cambridge, MA<br />
Prof. Dr. Siegfried Neumann (Industry)<br />
Fa. E. Merck<br />
Forschung DIAG<br />
Darmstadt<br />
Prof. Dr. Christiane Nüsslein-Volhard<br />
Max-Planck-Institut<br />
<strong>für</strong> Entwicklungsbiologie<br />
Tübingen<br />
Prof. Dr. Klaus Rajewsky<br />
Institut <strong>für</strong> Genetik<br />
Universität Köln<br />
Prof. Dr. Martin Schwab<br />
Institut <strong>für</strong> Hirnforschung<br />
Universität Zürich<br />
6 7
Vorbemerkungen des Direktors<br />
In den fünfzehn Jahren seines Bestehens hat das<br />
<strong>ZMBH</strong> gezeigt, daß es möglich ist, erfolgreich die<br />
Aufgaben zu übernehmen, gleichzeitig Forschung<br />
auf hochkompetitiven Gebieten <strong>der</strong> Molekular- und<br />
Zellbiologie zu betreiben, Studenten aus- und junge<br />
Wissenschaftler weiterzubilden und unabhängigen<br />
Nachwuchsgruppen jede För<strong>der</strong>ung zu geben. Als Universitätsinstitut<br />
gegründet, liegt <strong>der</strong> Schwerpunkt <strong>der</strong><br />
Aufgaben des <strong>ZMBH</strong> in <strong>der</strong> Verbindung von exzellenter<br />
Grundlagenforschung mit ebensolchen Lehrprogrammen<br />
<strong>für</strong> Studierende auf allen Stufen ihrer<br />
Ausbildung. Die Symbiose zwischen Forschung und<br />
Lehre, glaube ich heute feststellen zu können, haben<br />
wir früh erreicht. Bereits drei Jahre nach seiner Gründung<br />
führte das <strong>ZMBH</strong> ein eigenes <strong>für</strong> seine Diplomanden<br />
und Doktoranden obligatorisches Studienprogramm<br />
<strong>für</strong> Graduierte ein.<br />
Wer das Treppenhaus zu den Labors des <strong>ZMBH</strong><br />
betritt, wird an <strong>der</strong> Tür oftmals ein Poster finden, mit<br />
dem einer unserer Wissenschaftler einen Methodenkurs<br />
<strong>für</strong> unsere Graduierten ankündigt. Da das <strong>ZMBH</strong><br />
als Department eingerichtet wurde, in dem alle seine<br />
Einrichtungen von den Wissenschaftlern gleichberechtigt<br />
benutzt werden können, ist die Vermittlung von<br />
Technologien an unsere Studenten und Mitarbeiter<br />
eine unserer Aktivitäten, um <strong>der</strong> Herausfor<strong>der</strong>ung in<br />
<strong>der</strong> <strong>Biologie</strong>, Biomedizin und Biotechnologie beim<br />
Start in das neue Jahrtausend zu begegnen. Die Entwicklung<br />
<strong>der</strong> mo<strong>der</strong>nen <strong>Biologie</strong> hängt in steigendem<br />
Maße von Methoden ab und steht auch in steigendem<br />
Maße im Austausch mit <strong>der</strong> Biotechnologie und Medizin,<br />
nicht nur mit Bezug auf die Anwendung neuer<br />
grundlegen<strong>der</strong> Erkenntnisse, son<strong>der</strong>n auch mit neuen<br />
Methoden und Fragestellungen. Als Vorsitzen<strong>der</strong><br />
8<br />
des Vereins „BioRegion Rhein-Neckar-Dreieck, e.V.“<br />
erfahre ich beinahe täglich die Bedeutung dieses engen<br />
Dialogs, <strong>der</strong> <strong>für</strong> die För<strong>der</strong>ung des Transfers zwischen<br />
Grundlagenforschung und ihrer innovativen Anwendung<br />
in <strong>der</strong> Medizin und Biotechnologie notwendig<br />
ist.<br />
Die größte Herausfor<strong>der</strong>ung in <strong>der</strong> <strong>Biologie</strong> und Biomedizin<br />
besteht <strong>der</strong>zeit darin, daß die vollständigen<br />
Genomsequenzen von mehr als einem Dutzend Prokaryonten,<br />
von Hefe, von Caenorhabditis elegans und<br />
von Drosophila melanogaster aufgeklärt wurden, aber<br />
die darin enthaltene Information damit noch nicht<br />
dekodiert ist. Die vollständigen Genomsequenzen weiterer<br />
multizellulärer Eukaryonten, einschließlich <strong>der</strong><br />
des Menschen, werden bald bekannt sein. Der erste<br />
deutsche Beitrag, die vollständige Genomsequenz von<br />
Mycoplasma pneumoniae, kam übrigens von unserem<br />
Kollegen Richard Herrmann. Die Genomik wird von<br />
ihrer weitgehend deskriptiven Phase des Sequenzierens,<br />
die wohl nur noch fünf bis zehn Jahre andauern<br />
wird, zur nächsten Ebene des Verstehens, wie Gene<br />
funktionieren und zusammenwirken, übergehen. Hier<strong>für</strong><br />
müssen Methoden entwickelt werden, mit denen<br />
aufgeklärt werden kann, wie die im Genom kodierte<br />
Information in biologische Prozesse in Zellen, Organen<br />
und ganzen Organismen übersetzt wird. Um diesen<br />
Herausfor<strong>der</strong>ungen auf <strong>der</strong> Ebene <strong>der</strong> Technologien<br />
und Methodologien zu begegnen, wird das <strong>ZMBH</strong> als<br />
flexible Organisation seinen Wissenschaftlern wichtige<br />
neue Dienstleistungen in Form <strong>der</strong> bewährten<br />
Service-Einheiten zur Verfügung stellen. Gegenwärtig<br />
werden folgende neue Technologien und Methoden in<br />
unserem Institut etabliert: Hochleistungs-Mikroskopie<br />
als neue zentrale Einrichtung, Massenspektroskopie<br />
Introductory Remarks of the Director<br />
In the fifteen years of its existence, the <strong>ZMBH</strong> has<br />
proven that it is possible to successfully address the<br />
mission to conduct research in the forefront in highly<br />
competitive fields of molecular and cellular biology, to<br />
educate students, to train young scientists and to give<br />
full support to independent junior groups. Because it<br />
was designed as a university institute, the focus of the<br />
<strong>ZMBH</strong> is to combine basic research with excellent<br />
education programs for students at all levels. Three<br />
years after its inauguration, the <strong>ZMBH</strong> introduced its<br />
own graduate program obligatory for its diploma and<br />
doctoral students.<br />
If you enter the staircase leading to the laboratories,<br />
you may encounter at the door a poster advertising<br />
a practical course held for our graduate students by<br />
one of our faculty members. Because the <strong>ZMBH</strong> was<br />
designed as a department in which resources are open<br />
to the <strong>ZMBH</strong> faculty, transferring technology to our<br />
students and coworkers is just one of our activities to<br />
meet with the challenge in biology, biomedicine and<br />
biotechnology at the start of this new millennium. The<br />
development of mo<strong>der</strong>n biology increasingly depends<br />
on methods and is also increasingly in exchange with<br />
biotechnology and medicine, not only in regard to the<br />
application of discoveries in basic science but also<br />
by the provision of methods and scientific problems.<br />
As chairman of the BioRegion Rhein-Neckar-Triangle<br />
association, I experience almost daily the importance<br />
of having a close dialogue to exchange information<br />
between fundamental research and its innovative<br />
application in medicine and biotechnology industry.<br />
One of the major discoveries of the past years driving<br />
the progress in biology and biomedicine is the fact that<br />
we now know the complete genomic sequences for<br />
more than a dozen prokaryotic organisms as well as<br />
that of yeast, Caenorhabditis elegans and Drosophila<br />
melanogaster. The first contribution of Germany, the<br />
complete genomic sequence of Micrococcus pneumoniae,<br />
came from our colleague Richard Herrmann.<br />
The full genomic sequences of further multicellular<br />
eukaryotes, including humans will soon be known.<br />
Genomics will be passing from a largely descriptive<br />
phase of genomic sequencing, that may only last for<br />
another five to ten years, to the next level of un<strong>der</strong>standing<br />
of how genes function and interact. For this,<br />
methods need to be developed which allow to uncover<br />
how the information encoded in the genome is translated<br />
into biological processes within cells, organs and<br />
complete organisms. To meet the challenges at the<br />
level of technologies and methodologies, the <strong>ZMBH</strong><br />
as a flexible organization continues to provide to its<br />
faculty essential novel services through its powerful<br />
infrastructure of central service units. New technologies<br />
and methods are being presently set up at our<br />
institute: advanced light microscopy as a new central<br />
service unit, mass spectrometry for protein identification<br />
and sequencing within the existing biomolecular<br />
chemistry unit, advanced cell sorting for the selection<br />
and isolation of rare genotypes of transfected eukaryotic<br />
cells and a transgenic Drosophila service as part<br />
of our animal house facility. We are currently discussing<br />
the instalment of a DNA microarray technology<br />
service unit as part of biomolecular chemistry. Microarray<br />
technology that detects mRNA promises to measure<br />
expression of genomes in cells in response to<br />
internal or external signals and is therefore of interest<br />
to most of the <strong>ZMBH</strong> faculty. To meet with the laboratory<br />
space demand caused by the new central services,<br />
we will expand and build space for offices on top of<br />
9
zur Proteinidentifizierung und -sequenzierung in <strong>der</strong><br />
bestehenden Service-Einheit Biomolekulare Chemie,<br />
hochentwickeltes Cell Sorting zur Anreicherung und<br />
Isolierung seltener Genotypen in transfizierten Zellen,<br />
und <strong>der</strong> Service transgener Drosophila als Abteilung<br />
unserer Versuchstierhaltung. Aktuell diskutieren wir<br />
die Einrichtung <strong>der</strong> DNA-‘Microarray‘-Technologie<br />
als Teil unserer zentralen Einheit Biomolekulare<br />
Chemie. Mit <strong>der</strong> vielversprechenden ‚Microarray‘-<br />
Technologie zur Detektion von mRNA können die<br />
Expressionsmuster von Genomen in Zellen als Antwort<br />
auf interne und externe Signale gemessen werden.<br />
Dies ist von zentralem Interesse <strong>für</strong> die meisten Forschungsgruppen<br />
des <strong>ZMBH</strong>. Um dem Laborflächenbedarf<br />
<strong>der</strong> neuen zentralen Dienste zu entsprechen,<br />
werden wir das Institutsgebäude mit einem Dachpavillon<br />
aufstocken, um Platz <strong>für</strong> Büros zu schaffen. Das<br />
Büro des Direktors und solche zentralen Einheiten,<br />
die keine voll installierten Laborflächen brauchen, die<br />
bisher aber noch in solchen untergebracht sind (Biocomputing<br />
und Dokumentation), werden in das neue<br />
fünfte Stockwerk hinaufziehen - hoffentlich im Herbst<br />
nächsten Jahres. Das <strong>ZMBH</strong> ist glücklicherweise in<br />
<strong>der</strong> Lage, mit <strong>der</strong> Unterstützung von zwei seiner Professoren,<br />
den größten Teil <strong>der</strong> Baukosten mit Mitteln<br />
seines ‚overheads‘ zu finanzieren.<br />
In Hinsicht auf seine wissenschaftliche Produktivität,<br />
seine Lehraktivitäten und seine Berufungspolitik im<br />
Zeitraum dieses <strong>Bericht</strong>s hat das <strong>ZMBH</strong> bewiesen,<br />
daß es eine flexible Universitätseinrichtung mit kompromißlosen<br />
Qualitätsstandards ist, daß es fähig ist,<br />
aktuelle Forschungsthemen und Lehrinhalte zu implementieren.<br />
Das <strong>ZMBH</strong> als ‚center of exzellence‘ kann<br />
sicherlich auch als eine universitäre Modelleinrichtung<br />
bezeichnet werden, das in <strong>der</strong> Lage ist, den Herausfor<strong>der</strong>ungen<br />
<strong>der</strong> mo<strong>der</strong>nen <strong>Biologie</strong> wissenschaftlich<br />
zu entgegnen. Bisherige Bilanz:<br />
10<br />
Wissenschaftliche Leistungen – wie von unseren<br />
Forschungsgruppenleitern in diesem und den vorausgegangenen<br />
Jahresberichten dargestellt, waren viele<br />
Höhepunkte und wirkliche Durchbrüche zu verzeichnen.<br />
Für mich wurden die bemerkenswertesten erreicht<br />
auf den Gebieten <strong>der</strong> Kontrolle <strong>der</strong> eukaryontischen<br />
Genexpression unter physiologischen und artifiziellen<br />
Bedingungen (Tetrazyklin-System), <strong>der</strong> Chromatin<br />
induzierten epigenetischen Kontrolle <strong>der</strong> Genexpression<br />
in Drosophila und Mammalia, des Targeting und<br />
innerzellulären Sortings von Molekülen einschließlich<br />
des Transports zwischen Kern und Cytoplasma, <strong>der</strong><br />
Ubiquitin-abhängigen Proteolyse, <strong>der</strong> Funktion und<br />
Regulation von Chaperonen, <strong>der</strong> Neurobiologie <strong>der</strong><br />
erregenden Neurotransmission, <strong>der</strong> Myelin-Genetik<br />
und <strong>der</strong> Neurodegeneration sowie <strong>der</strong> Wirt-Parasiten/<br />
Virus Interaktionen.<br />
Modell <strong>ZMBH</strong> – mit dem Einführen neuer Strukturen<br />
<strong>für</strong> ein Universitätsinstitut vor 15 Jahren – in dem<br />
Grundlagenforschung in kleinen Gruppen betrieben<br />
wird, die effizient durch eine Infrastruktur gemeinsam<br />
genutzter leistungsfähiger wissenschaftlicher, technischer<br />
und administrativer Dienste unterstützt werden<br />
(rund 50% unserer Finanzmittel vom Land Baden-<br />
Württemberg werden hier<strong>für</strong> verwendet und nur die<br />
an<strong>der</strong>e Hälfte dieser Mittel wird den einzelnen Forschungsgruppen<br />
direkt zur Verfügung gestellt) – jetzt<br />
übernommen von einem <strong>Zentrum</strong> <strong>der</strong> Universität<br />
Tübingen und zwei neuen Zentren in Heidelberg<br />
Umfassende Reform des Curriculums <strong>der</strong> Fakultät<br />
<strong>für</strong> <strong>Biologie</strong>, die vom <strong>ZMBH</strong> initiiert und entwickelt<br />
wurde, und an die sich jetzt eine erfolgreiche Initiative<br />
zur Einführung eines internationalen Bachelor- und<br />
Master-Studienganges <strong>für</strong> <strong>Molekulare</strong> und Zellbiologie<br />
anschließt<br />
Hervorragende Ausbildung und Unterstützung<br />
unserer Diplomanden, Doktoranden, Postdocs, For-<br />
the <strong>ZMBH</strong>. The office of the director and of those<br />
central units that do not need but presently use fully<br />
equipped lab space (biocomputing and documentation)<br />
will be moved to the new fifth floor, hopefully by<br />
next fall. The <strong>ZMBH</strong> is fortunate to be able to cover<br />
most of the building costs using its overhead money<br />
and with the financial help of two senior faculty members.<br />
Regarding its scientific output, teaching activities and<br />
hiring policy for the period covered by this report, the<br />
<strong>ZMBH</strong> has proven to be a flexible university institution<br />
with uncompromising standards of quality, continuing<br />
adaptation to changing needs and international<br />
recognition as a center of excellence and for its added<br />
value that include:<br />
Scientific achievements, as documented by the<br />
research reports of our group lea<strong>der</strong>s in this issue<br />
and previous reports, we had many highlights and<br />
real break-throughs. For me, the most notable being<br />
achieved in the field of control of eucaryotic gene<br />
expression un<strong>der</strong> physiological and artificial conditions<br />
(tetracyclin system), of chromatin-induced epigenetic<br />
control of gene expression in Drosophila and<br />
mammals, protein targeting and intracellular sorting<br />
including transport between nucleus and cytoplasm,<br />
ubiquitin-dependent proteolysis, function and regulation<br />
of chaperons, neurobiology of excitatory neurotransmission,<br />
myelin genetics and neurodegeneration<br />
and of host-parasite/virus interactions.<br />
Conceptual lea<strong>der</strong>ship in introducing a new structure<br />
for an university institute 15 years ago - in which the<br />
research done by small groups is efficiently supported<br />
by an efficient infrastructure with jointly used scientific,<br />
technical and administrative service (approximately<br />
50% of our funds from the State of Baden-<br />
Wuerttemberg is allocated to it and only the other half<br />
of these funds is passed on directly to the individual<br />
groups) - now adopted by one center at the University<br />
of Tuebingen and two new centers in Heidelberg<br />
Initiation of a distinct, revised curriculum of the<br />
faculty for biology, conceived by the <strong>ZMBH</strong> is now<br />
being successfully followed by plans to inaugurate an<br />
international curriculum of a bachelors and masters<br />
program in Molecular and Cellular Biology<br />
Outstanding training and career possibilities of<br />
diploma students, doctoral students, and postdoctoral<br />
fellows, group lea<strong>der</strong>s and visitors – alone five former<br />
group lea<strong>der</strong>s of the <strong>ZMBH</strong> have moved to Max-<br />
Planck-Institutes as directors<br />
The period of 1998 – <strong>2000</strong><br />
In the past 32 months the <strong>ZMBH</strong> has seen an unprecedented<br />
turnover among its group lea<strong>der</strong>s. Stefan<br />
Jentsch and Klaus Nave moved to Max-Planck-Institutes<br />
in Munich and Goettingen, respectively. Our<br />
“founding members” Hermann Bujard and Heinz<br />
Schaller reached the official retirement age and are<br />
now no longer allowed to sign official documents, a<br />
discrimination that has been overcome some time ago<br />
in other countries. As documented by their reports,<br />
Hermann Bujard and Heinz Schaller have both been<br />
extremely successful with their projects and publications.<br />
We highly appreciate that both scientists continue<br />
doing their research and teaching at the <strong>ZMBH</strong><br />
as usual. To enable this, we changed our bylaws and<br />
created what we call the “Emeritus Regelung”, which<br />
allows retired group lea<strong>der</strong>s to continue research as<br />
long as they obtain external funding and their projects<br />
are accepted by the <strong>ZMBH</strong>’s scientific advisory board<br />
and faculty. Contracts are for one to three years and<br />
renewable.<br />
Another pleasant development is that Renato Paro was<br />
11
schungsgruppenleiter und Gastwissenschaftler - allein<br />
fünf frühere Gruppenleiter des <strong>ZMBH</strong> sind als Direktoren<br />
an Max-Planck-Institute übergewechselt.<br />
Zum <strong>Bericht</strong>szeitraum 1998 - <strong>2000</strong><br />
In den vergangenen 32 Monaten sah das <strong>ZMBH</strong> einen<br />
beispiellosen Personalwechsel bei seinen Gruppenleitern.<br />
Stefan Jentsch und Klaus-Armin Nave wechselten<br />
zu Max-Planck-Instituten in München und Göttingen.<br />
Unsere „Gründungsmitglie<strong>der</strong>“ Hermann Bujard<br />
und Heinz Schaller erreichten das offizielle Pensionsalter<br />
und dürfen jetzt keine offiziellen Dokumente mehr<br />
unterzeichnen, eine Diskriminierung, die in an<strong>der</strong>en<br />
Län<strong>der</strong>n vor einiger Zeit abgeschafft wurde. Wie in<br />
ihren Forschungsberichten dokumentiert, waren Hermann<br />
Bujard und Heinz Schaller beide weiterhin<br />
außerordentlich erfolgreich mit ihren Forschungsprojekten<br />
und Veröffentlichungen. Wir schätzen es<br />
sehr, daß Beide ohne weitere Einschränkungen ihre<br />
Forschung und Lehre am <strong>ZMBH</strong> fortsetzen. Hier<strong>für</strong><br />
haben wir unsere Institutssatzung geän<strong>der</strong>t und eine<br />
spezielle, wie wir sie nennen „Emeritus-Regelung“<br />
geschaffen. Letztere ermöglicht es Gruppenleitern im<br />
Ruhestand, ihre Forschungsarbeiten weiterzuführen,<br />
solange sie hier<strong>für</strong> Drittmittel einwerben können und<br />
vom Wissenschaftlichen Beirat und vom Kollegium<br />
des <strong>ZMBH</strong> hier<strong>für</strong> die Zustimmung erhalten. Diese<br />
wird befristet <strong>für</strong> 1 - 3 Jahre gegeben und ist verlängerbar.<br />
Eine zukunftsweisende erfreuliche Entwicklung wurde<br />
vergangenen Dezember mit <strong>der</strong> Berufung von Renato<br />
Paro als Nachfolger von Stefan Jentsch eingeleitet.<br />
Zum Zeitpunkt, an dem dieser <strong>Bericht</strong> geschrieben<br />
wird, haben wir noch drei freie Professuren. Wir rechnen<br />
damit, daß zwei dieser Professuren noch im Jahr<br />
<strong>2000</strong> wie<strong>der</strong>besetzt werden, und wir sind optimistisch,<br />
daß die Dritte kurz darauf besetzt werden kann.<br />
12<br />
Im Jahre <strong>2000</strong> ist Blanche Schwappach von <strong>der</strong> Universität<br />
Californien, San Francisco einem Ruf als neue<br />
Nachwuchsgruppenleiterin ans <strong>ZMBH</strong> gefolgt. Ihre<br />
innovativen Arbeiten über ER-Trafficking-Signale, die<br />
eine essentielle Kontrollfunktion <strong>für</strong> ein Plasmamembran-Kanalprotein<br />
haben, überzeugten uns bei <strong>der</strong> Auswahl<br />
unter mehr als 50 Bewerbern. Blanche Schwappachs<br />
Forschungsthematik auf dem Gebiet <strong>der</strong> Regulation<br />
des intrazellulären Transports paßt gut in unser<br />
Forschungsprogramm. Wir sind davon überzeugt, daß<br />
ihre experimentellen Strategien von unserer Expertise<br />
im Bereich <strong>der</strong> molekularen Zellbiologie profitieren<br />
werden und daß wir ihre Expertise bei <strong>der</strong> Anwendung<br />
ausgefeilter Methoden des ‚Cell Sortings‘ zur Selektion<br />
interessanter, selten vorkommen<strong>der</strong> Genotypen/<br />
Phenotypen transfizierter eukaryontischer Zellen gut<br />
nutzen werden.<br />
appointed as successor of Stefan Jentsch last December.<br />
At the time of writing, we have three open tenure<br />
positions. We expect two of the three positions to be<br />
filled during <strong>2000</strong> and we are optimistic that the third<br />
professorship will follow soon after.<br />
In July <strong>2000</strong>, we hired Blanche Schwappach from the<br />
University of California, San Francisco as a new junior<br />
group lea<strong>der</strong>. Her innovative studies of ER trafficking<br />
signals serving an essential quality control function<br />
for a plasma membrane channel protein made us select<br />
her from over 50 applicants. Blanche Schwappach’s<br />
research focus on the regulation of intracellular transport<br />
mechanism fits well into our research program.<br />
We are confident that her experimental strategies will<br />
benefit from our expertise in molecular cell biology,<br />
and that we will benefit from her expertise in using<br />
sophisticated cell sorting for the selection of interesting<br />
rare genotypes/phenotypes of transfected eukaryotic<br />
cells.<br />
Due to their outstanding achievements in research and<br />
teaching, Gerd Multhaup and Percy Knolle were promoted<br />
to “project group lea<strong>der</strong>”. The host for both<br />
is currently the director. The status of “project group<br />
lea<strong>der</strong>” at the <strong>ZMBH</strong> as anchored to our bylaws,<br />
allowing the <strong>ZMBH</strong> to advance successful young scientists<br />
to formal independence, when justified by their<br />
achievements. Further requirements are independent<br />
research, grant support, the German “habilitation” and<br />
formal approval by the <strong>ZMBH</strong>‘s faculty. The appointed<br />
project group lea<strong>der</strong> receives lab space and financial<br />
support from our state budget but stays associated with<br />
one of the permanent groups for organizational purposes.<br />
Consi<strong>der</strong>able financial investments have again been<br />
allocated to our infrastructure. We are proud of the<br />
achievements of our transgenic unit un<strong>der</strong> its experienced<br />
and skilled head Juergen Weiss. The remarkable<br />
success of Frank Zimmermann and Domenico Basta to<br />
produce transgenic foun<strong>der</strong>s is acknowledged not only<br />
in the <strong>ZMBH</strong>. The latest addition to the animal facility<br />
is the service for transgenic flies that was installed<br />
to meet the increasing demands of several groups. The<br />
biomedical chemistry unit received a new Q-TOF mass<br />
spectrometer suited for proteomics from the Deutsche<br />
Forschungsgemeinschaft. We are indebted to Richard<br />
Herrmann for acting as interim head of this unit<br />
after Rainer Frank‘s departure. Thomas Ruppert will<br />
become the new head effective October <strong>2000</strong>. He is<br />
experienced in proteomics and worked previously at<br />
the Institute for Biochemistry, Charité, Berlin. The<br />
unit for high-resolution microscopy is currently being<br />
set up on the third floor with the assistance of Axel<br />
Baumm. It will be moved to laboratories on the first<br />
floor, currently being used as the director‘s office as<br />
13
In Anerkennung ihrer hervorragenden Forschung und<br />
Lehre wurden Gerd Multhaup und Percy Knolle zum<br />
„Projektgruppenleiter“ ernannt. Der ‚host‘ <strong>für</strong> beide<br />
ist gegenwärtig <strong>der</strong> Direktor. Der Status eines „Projektgruppenleiters“,<br />
wie er in den Institutsregelungen<br />
festgelegt ist, erlaubt es dem <strong>ZMBH</strong>, erfolgreichen<br />
jungen Wissenschaftlern formale Unabhängigkeit zu<br />
geben, wenn <strong>der</strong> Fortschritt ihrer wissenschaftlichen<br />
Arbeiten dies zuläßt. Weitere Voraussetzungen sind<br />
unabhängige Forschungsvorhaben, projektbezogene<br />
Drittmittel, die Habilitation und die formelle Zustimmung<br />
des <strong>ZMBH</strong>-Kollegiums. Der ernannte Projektgruppenleiter<br />
erhält eigenen Laborraum und eigene<br />
Institutsmittel, bleibt aus organisatorischen Gründen<br />
aber mit einer <strong>der</strong> permanenten Gruppen assoziiert.<br />
Erhebliche Investitionsmittel wurden wie<strong>der</strong> <strong>für</strong> unsere<br />
Infrastruktur aufgewendet. Wir sind stolz auf die<br />
exzellenten Leistungen unserer Transgenen Einheit<br />
unter <strong>der</strong> erfahrenen und fachmännischen Leitung von<br />
Jürgen Weiß. Die bemerkenswerten Erfolge von Frank<br />
Zimmermann und Domenico Basta bei <strong>der</strong> Herstellung<br />
transgener Stämme werden nicht nur vom <strong>ZMBH</strong><br />
hoch geschätzt. Die jüngste Erweiterung <strong>der</strong> Versuchstierhaltung<br />
ist <strong>der</strong> Service <strong>für</strong> transgene Fliegen, <strong>der</strong><br />
etabliert wurde, um dem anwachsenden Bedarf mehrerer<br />
Gruppen nachzukommen. Die Einheit Biomolekulare<br />
Chemie erhielt von <strong>der</strong> Deutschen Forschungsgemeinschaft<br />
ein spezielles Q-TOF-Massenspektrometer<br />
<strong>für</strong> die Proteomik. Wir sind Richard Herrmann dankbar,<br />
daß er die zentrale Einheit vertretungsweise leitet,<br />
seit Rainer Frank ausgeschieden ist. Thomas Ruppert<br />
wird im Oktober <strong>2000</strong> die Leitung übernehmen; er hat<br />
zuvor am Institut <strong>für</strong> Biochemie, Charité, Berlin gearbeitet<br />
und bringt große Erfahrung im Bereich <strong>der</strong> Proteomik<br />
mit. Die Einheit <strong>für</strong> hochauflösende Mikroskopie<br />
wird jetzt zunächst im 3. Stockwerk mit <strong>der</strong><br />
Assistenz von Axel Baumm eingerichtet. Sie wird in<br />
14<br />
Laborräume im 1. Stockwerk, die zur Zeit noch als<br />
Sekretariat des Direktors genutzt werden, umziehen,<br />
sobald dieses in den neuen Dachpavillon einziehen<br />
kann. Wie zuvor bemerkt, sind weitere Service-Einheiten<br />
<strong>für</strong> DNA-‘Microarray‘-Technologie und <strong>für</strong> quantitative<br />
PCR geplant. Der Leiter <strong>der</strong> Verwaltung, Jürgen<br />
Auer und sein Team haben erfolgreich ein den spezifischen<br />
Anfor<strong>der</strong>ungen <strong>der</strong> Buchhaltung des <strong>ZMBH</strong><br />
angepaßtes EDV-Programm eingeführt. Hiermit konnte<br />
die Effizienz und Geschwindigkeit <strong>der</strong> Verwaltungsdienste<br />
weiter erhöht werden. Ohne die EDV-Abteilung<br />
unter <strong>der</strong> Leitung von Raphael Mosbach gäbe es<br />
keine effektive und aktuelle Bioinformatik im <strong>ZMBH</strong>.<br />
In diesem Zusammenhang sind mit Anerkennung die<br />
essentielle Zuarbeit unserer von Matthias Pawlitschko<br />
und Gert Stegmüller geleiteten Elektro- und Mechanikwerkstätten<br />
aufzuführen. Die hohe Priorität, die<br />
das Biocomputing bei uns hat, erlaubt es dem <strong>ZMBH</strong><br />
jetzt, mit einem Lehrprogramm <strong>für</strong> Bioinformatik zu<br />
beginnen, das unsere Studenten in die „Exploration“<br />
von Datenbanken einführt, um Struktur und Funktion<br />
aus Sequenzen vorherzusagen, o<strong>der</strong> Einsicht in die<br />
Evolution von intra- und interspezies Sequenzvariationen<br />
und ihren Folgen zu gewinnen. Die exzellenten<br />
graphischen Arbeiten von Yves Cully, von <strong>der</strong> Dokumentationsabteilung<br />
bereitgestellt, haben entscheidend<br />
zum Erfolg bei vielen Vorträgen und bei <strong>der</strong> Präsentation<br />
von Postern durch <strong>ZMBH</strong>-Mitglie<strong>der</strong> auf<br />
Tagungen und Ausstellungen beigetragen. Abschließend<br />
möchte ich Frau Gerlind Güth-Köhler, Frau Heidemarie<br />
Demuth und Dr. Hans Peter Blaschkowski <strong>für</strong><br />
ihre effiziente und kreative Unterstützung des Direktors<br />
danken.<br />
In diesem Jahr haben unsere Labors im 1. Stockwerk<br />
eine Klimaanlage bekommen. Diejenigen von uns, die<br />
auf diesem Stock arbeiten, können endlich auch im<br />
Sommer mit Drosophila experimentieren und müssen<br />
soon as the office can be moved to the new fifth floor.<br />
As already indicated, services for DNA chip technology<br />
and quantitative PCR are in the planning stage.<br />
The head of the administration Juergen Auer and his<br />
team have successfully implemented software programs<br />
tailored for the special needs of bookkeeping at<br />
the <strong>ZMBH</strong>. This has further increased the efficiency<br />
and speed of our administrative support. The biocomputing<br />
unit headed by Raphael Mosbach constantly<br />
increases our computing power and network support.<br />
In this context, we acknowledge the essential support<br />
of our electrical and mechanical workshop led by Matthias<br />
Pawlitschko and Gert Stegmueller. With the hardware<br />
support of biocomputing at the <strong>ZMBH</strong> a teaching<br />
program in bioinformatics can now be launched<br />
this year to prepare our students for the “mining” of<br />
databases and for predicting both structure and function<br />
from sequence, as well as evolution of intra- and<br />
interspecies sequence variation and its consequences.<br />
The artwork provided by Yves Cully of our documentation<br />
service has been critical for the success of<br />
the many talks, presentations of posters presented by<br />
<strong>ZMBH</strong> members at meetings and exhibitions. Last<br />
but not least, I am grateful to Gerlind Gueth-Koehler,<br />
Heidemarie Demuth and Dr. Hans Peter Blaschkowski<br />
for their very efficient and creative support of the<br />
director.<br />
This year our laboratories of the first floor have<br />
received air-conditioning. Those of us working in this<br />
floor can now continue with our experiments using<br />
Drosophila as a model during summer time and do not<br />
have to worry any longer about losing valuable strains.<br />
We very much hope that the laboratories in the other<br />
floors will receive the same air-conditioning standard<br />
in the coming two years. To meet with the or<strong>der</strong>s of the<br />
fire brigade and the very limited lab space allocated to<br />
each research group, the <strong>ZMBH</strong> had to invest in freez-<br />
ers, suitable for using in the floors connecting the laboratories.<br />
This investment was again made possible by<br />
the overhead brought in by those groups having EC,<br />
HFSP and industrial support.<br />
After fourteen years of provisional delivery and waste<br />
management, the <strong>ZMBH</strong> received a new building for<br />
that purpose. We are grateful to the University for the<br />
improvement. In or<strong>der</strong> to make life easier for our busy<br />
caretaker Michael Konrad, we hope that the waste<br />
containers will soon be moved into this shelter.<br />
The <strong>ZMBH</strong> itself shall become a building site this fall.<br />
As mentioned earlier, we are looking forward to the<br />
new office space on the fifth floor. If everything works<br />
according to plan, we shall move into the new offices<br />
next fall and be able to set free the desperately needed<br />
lab space for our new service units.<br />
We very much regret that the new building for physics<br />
which is presently un<strong>der</strong> construction next to<br />
State Minister von Trotha visits the mechanical workshop<br />
of the <strong>ZMBH</strong>.<br />
15
nicht mehr den Verlust wichtiger Stämme be<strong>für</strong>chten.<br />
Wir hoffen jetzt sehr, daß auch die Labors in den an<strong>der</strong>en<br />
Stockwerken den selben Klimatisierungs-Standard<br />
in den nächsten Jahren bekommen. Um den Anordnungen<br />
<strong>der</strong> Feuerpolizei nachzukommen und um die<br />
knappen Laborflächen, die je<strong>der</strong> Forschungsgruppe<br />
zur Verfügung stehen, optimal zu nutzen, mußte das<br />
<strong>ZMBH</strong> umfangreiche Investitionen in Spezialkühlschränke<br />
tätigen, die in den Verbindungsfluren zwischen<br />
den Labors aufgestellt werden dürfen. Diese<br />
Investitionen werden wie<strong>der</strong>um durch den ‚overhead‘<br />
von den Gruppen ermöglicht, die über Drittmittel <strong>der</strong><br />
EU, des HFSP und <strong>der</strong> Industrie verfügen.<br />
Nach vierzehn Jahren Improvisation bei <strong>der</strong> Lagerhaltung<br />
und dem Abfall-Management hat das <strong>ZMBH</strong><br />
einen Anbau <strong>für</strong> diese Zwecke erhalten. Um die Arbeit<br />
unseres eifrigen Hausmeisters Michael Konrad zu<br />
erleichtern, hoffen wir, daß die Abfallcontainer hier<br />
endlich ordnungsgemäß untergebracht werden.<br />
Das <strong>ZMBH</strong>-Gebäude selbst wird in diesem Herbst<br />
eine Baustelle werden. Wie zuvor geschil<strong>der</strong>t, sehen<br />
wir erwartungsvoll dem Dachpavillon entgegen. Wenn<br />
alles nach Plan verläuft, werden wir die neuen Büroräume<br />
im nächsten Jahr beziehen und damit die Laborflächen<br />
räumen, die dringend <strong>für</strong> unsere neuen Service-Einheiten<br />
benötigt werden.<br />
Wir bedauern außerordentlich, daß das neue Institut<br />
<strong>für</strong> Physik, das jetzt direkt neben dem <strong>ZMBH</strong> errichtet<br />
wird, uns sein Technikum zuwendet und nicht, wie<br />
wir wünschten, seine Hörsäle. Der einzige Kompromiß,<br />
den wir in Verhandlungen erreichen konnten, war<br />
ein größerer Abstand zwischen dem Physik-Technikum<br />
und dem <strong>ZMBH</strong>. Nach den ursprünglichen Planungen<br />
wäre <strong>der</strong> Physik-Neubau so nahe an unser Institut<br />
herangesetzt worden, daß wir erhebliche Störungen<br />
bei unseren erschütterungsempfindlichen mikroskopischen<br />
Untersuchungen be<strong>für</strong>chten mußten.<br />
16<br />
Vor vier Jahren wurde das eigene Studienprogramm<br />
<strong>für</strong> Doktoranden am <strong>ZMBH</strong> durch zwei „Graduierten-Kollegs“<br />
<strong>der</strong> Deutschen Forschungsgemeinschaft<br />
erweitert. Wir sind stolz darauf, daß diese „Graduierten-Kollegs“<br />
verlängert und weiterhin von Christine<br />
Clayton und Bernhard Dobberstein organisiert<br />
werden.<br />
Zur speziellen und flexiblen För<strong>der</strong>ung von Postdocs<br />
und Gastwissenschaftlern hat das <strong>ZMBH</strong>-Kollegium<br />
vor vier Jahren das „<strong>ZMBH</strong>-Stipendium“ gestiftet, das<br />
inzwischen an mehr als ein Dutzend junger Mitarbeiter<br />
und Gastwissenschaftler vergeben wurde.<br />
Die Tradition des <strong>ZMBH</strong>, wissenschaftlichen Austausch<br />
zu för<strong>der</strong>n, wurde im vergangenen Zeitraum<br />
ebenfalls weiterverfolgt. Im Jahr 1998 fand das<br />
<strong>ZMBH</strong>-Forum ‚Genetic Basis of Brain Function‘ statt,<br />
es wurde von Klaus-Armin Nave organisiert. Das<br />
<strong>ZMBH</strong>-Forum 1999 mit dem Titel ‚Pathogenetic Protozoa:<br />
Molecules, Structures & Mechanims‘ organisierte<br />
Christine Clayton. Beide Foren waren mit exzellenten<br />
Rednern besetzt und wie<strong>der</strong> sehr gut besucht.<br />
Heidelberg ist das Herz <strong>der</strong> BioRegion Rhein-Neckar-<br />
Dreieck. Aus einem Wettbewerb mit 17 an<strong>der</strong>en deutschen<br />
Regionen vor vier Jahren ging die BioRegion<br />
Rhein-Neckar-Dreieck als eine <strong>der</strong> drei Gewinner<br />
hervor. Der größte Teil <strong>der</strong> 50 Mio. DM, die bei diesem<br />
Wettbewerb erhalten wurden, wurde jetzt <strong>für</strong> Projekte<br />
15 junger Biotechnologie-Firmen vergeben. Der Beitrag<br />
des <strong>ZMBH</strong> wird durch die Tatsache unterstrichen,<br />
daß Bernhard Dobberstein bei <strong>der</strong> Gründung <strong>der</strong><br />
BioRegion den Wissenschaftsbereich koordinierte und<br />
daß ich zur Zeit <strong>der</strong> Vorsitzende des neu gegründeten<br />
Vereins „BioRegion Rhein-Neckar-Dreieck, e.V.“ bin.<br />
Neben den vielen günstigen Ereignissen <strong>der</strong> letzten<br />
Jahre und <strong>der</strong> fortlaufenden Unterstützung durch<br />
Rektor, Kanzler und Kanzlerin <strong>der</strong> Universität und<br />
the <strong>ZMBH</strong> faces us with its workshop and not, as<br />
we wished, with its lecture theater. The only compromise<br />
that we were able to negotiate was the distance<br />
between the physics workshop and the <strong>ZMBH</strong>. The<br />
original plan would have placed the two buildings in<br />
such a close proximity that we expected to encounter<br />
severe problems with our vibration sensitive microscopic<br />
studies.<br />
Four years ago the <strong>ZMBH</strong>‘s own study program for<br />
doctoral students was extended by two “Graduierten<br />
Kollegs” of the Deutsche Forschungsgemeinschaft.<br />
We are proud that these “Graduierten Kollegs” were<br />
renewed and continue to be organized by Christine<br />
Clayton and Bernhard Dobberstein.<br />
To permit special and flexible support for postdoctoral<br />
fellows and visitors the <strong>ZMBH</strong> faculty implemented<br />
the “<strong>ZMBH</strong> fellowship” four years ago which<br />
has meanwhile been awarded to more than a dozen fellows.<br />
The tradition at the <strong>ZMBH</strong> to promote scientific<br />
exchange was also continued in the recent past. In<br />
1998 the <strong>ZMBH</strong> Forum on “Genetic Basis of Brain<br />
Function” was organized by Klaus-Armin Nave. In<br />
the following year 1999, Christine Clayton organized<br />
the <strong>ZMBH</strong> Forum un<strong>der</strong> the theme “Pathogenic Protozoa:<br />
Molecules, Structures & Mechanisms”. Both<br />
were again best-received and well attended <strong>ZMBH</strong><br />
meetings.<br />
Heidelberg is in the heart of the BioRegion Rhine-<br />
Neckar-Triangle. Two years ago in competition with 17<br />
other German regions the BioRegion Rhine-Neckar-<br />
Triangle emerged as one of the three winners. The<br />
major part of the 50 million DM received from this<br />
competition has now been awarded to projects in fifteen<br />
start-up biotech companies. The contribution of<br />
the <strong>ZMBH</strong> is un<strong>der</strong>lined by the fact that Bernhard<br />
Dobberstein initially coordinated the scientific part of<br />
the application and I am presently chairman of the<br />
newly found BioRegion Rhine-Neckar-Triangle association.<br />
MdL Pfisterer und MdB Lamers (1. und 2. von links)<br />
besichtigen gentechnische Labors des <strong>ZMBH</strong>.<br />
Besides many favorable events from the last years and<br />
the continuing support by the Rector and Chancellor<br />
of the University and Minister von Trotha, there are<br />
developments which make us worry. Of major concern<br />
for us is that the position for a successor of Hermann<br />
Bujard is still pending. Furthermore, given the high<br />
turnover rate of the <strong>ZMBH</strong> faculty members, the time<br />
it takes to fill a tenured position, at present up to two<br />
years, is much too long in a highly competitive situation.<br />
In contrast, it takes the <strong>ZMBH</strong> only six to nine<br />
months from the advertisement to the start of a new<br />
junior group lea<strong>der</strong>. This is not due to a less careful<br />
selection process. The selection of a junior group<br />
lea<strong>der</strong> by the <strong>ZMBH</strong> faculty is done according to international<br />
academic standards and by seeking advice<br />
from our scientific advisory board.<br />
As repeatedly mentioned by the former director and<br />
cofoun<strong>der</strong> of the <strong>ZMBH</strong> Hermann Bujard we must<br />
17
durch Minister von Trotha, gibt es einige Sorge bereitende<br />
Entwicklungen. Mit wachsen<strong>der</strong> Beunruhigung<br />
sehen wir, daß die Stelle <strong>für</strong> den Nachfolger von Hermann<br />
Bujard immer noch nicht bewilligt ist. Außerdem<br />
ist, in Anbetracht <strong>der</strong> fortdauernd hochkompetitiven<br />
Personalsituation im Bereich <strong>der</strong> molekularen<br />
Biowissenschaften und bei dem gegebenen großen<br />
Wechsel <strong>der</strong> Mitglie<strong>der</strong> des <strong>ZMBH</strong>-Kollegiums, <strong>der</strong><br />
Zeitraum von gegenwärtig zwei Jahren <strong>für</strong> die Wie<strong>der</strong>besetzung<br />
frei gewordener Professuren viel zu groß.<br />
Das dies auch an<strong>der</strong>s geht, zeigen die Besetzungen<br />
von Stellen <strong>für</strong> Nachwuchsgruppenleiter. Das <strong>ZMBH</strong><br />
benötigt lediglich sechs bis neun Monate von <strong>der</strong> Ausschreibung<br />
einer freien Stelle bis zum Arbeitsbeginn<br />
eines neuen Nachwuchsgruppenleiters. Dies hat seinen<br />
Grund nicht in einem weniger aufwendigen Auswahlverfahren.<br />
Die Berufung eines Nachwuchsgruppenleiters<br />
durch das <strong>ZMBH</strong> wird internationalen akademischen<br />
Standards entsprechend und mit Beteiligung<br />
unseres Wissenschaftlichen Beirats durchgeführt.<br />
Wie wie<strong>der</strong>holt von dem früheren Direktor und Mitgrün<strong>der</strong><br />
des <strong>ZMBH</strong> Hermann Bujard dargelegt, müssen<br />
wir nachdrücklich darauf bestehen, daß die Institute<br />
<strong>der</strong> Universitäten in die Lage versetzt werden, die<br />
wichtigsten Orte <strong>der</strong> Forschung zu sein. Hier, an diesen<br />
Institutionen erhält die große Mehrzahl <strong>der</strong> jungen<br />
Wissenschaftler ihre erste und damit wegweisende<br />
Ausbildung, und die Qualität dieser Ausbildung wird<br />
durch die Qualifikation ihrer Lehrer bestimmt. Deshalb<br />
muß die Universität ihre Attraktivität <strong>für</strong> die Besten<br />
ihres Faches behalten und darf sie nicht an außenstehende<br />
Einrichtungen verlieren. Wir sind Minister von<br />
Trotha außerordentlich dankbar <strong>für</strong> die offene Diskussion<br />
unserer Besorgnis, das dieser essentielle Synergismus<br />
höchst gefährdet ist und hoffen, daß das neue<br />
Universitätsgesetz zu Än<strong>der</strong>ungen in die richtige Richtung<br />
führt. In einer Zeit, in <strong>der</strong> wir die höchst erfreu-<br />
18<br />
liche Zunahme <strong>der</strong> Zahl <strong>der</strong> biotechnischen Grün<strong>der</strong>firmen<br />
vor uns sehen, müssen wir realisieren, daß die<br />
Zukunft dieser Unternehmen von breitester Grundlagenforschung<br />
und einer großen Zahl bestens ausgebildeter<br />
Graduierter und promovierter Wissenschaftler<br />
abhängt. Beide Ziele können nur mit gut ausgestatteten<br />
und hochflexiblen Universitätseinrichtungen<br />
Minister von Trotha im Lehrlabor des <strong>ZMBH</strong>.<br />
erreicht werden. Es bleibt darauf hinzuweisen, daß<br />
die biomedizinische Forschung in Deutschland wie<strong>der</strong><br />
hinter die <strong>der</strong> USA zurückfällt, wenn die beteiligten<br />
Institutionen keine entsprechende Unterstützung<br />
bekommen. Daß die wissenschaftliche Grundlagenforschung<br />
an den Universitäten höhere Priorität bekommen<br />
muß, als ihr heute eingeräumt wird, veranlaßte<br />
das <strong>ZMBH</strong> konsequenterweise, einen weitgreifenden<br />
Dialog mit <strong>der</strong> Politik und Öffentlichkeit zu führen.<br />
Wir waren dankbar, daß <strong>der</strong> Ministerpräsident von<br />
Baden-Württemberg Erwin Teufel unsere Einladung<br />
annahm, anläßlich seiner Bereisung <strong>der</strong> BioRegion und<br />
des Technologieparks Heidelberg am 21. Januar 1999<br />
auch das <strong>ZMBH</strong> zu besuchen. Der Wirtschaftsminister<br />
des Landes Walter Döring besuchte das <strong>ZMBH</strong> am<br />
7. April 1998, um mit uns Aspekte des Technologie-<br />
stress and repeat that university institutes need to be<br />
able to maintain the essential sites for research. It is at<br />
these institutions where the vast majority of young scientists<br />
receive their first and thus life-deciding train-<br />
Fotis C. Kafatos, Director-General at EMBL, acknowledged<br />
Hermann Bujard's leading role in bringing the<br />
EMBL to Heidelberg (celebration of H. Bujard's 65th<br />
birthday).<br />
ing with the level of training being determined by the<br />
quality of teachers. Hence the university must hold<br />
attraction for the best people in a field and is not to<br />
lose them to outside institutions. We are most grateful<br />
to Minister von Trotha for the open discussion of our<br />
concerns and hope that the new Hochschulgesetz will<br />
lead to changes in the right direction. At a time where<br />
we witness an unprecedented increase in the number<br />
of biotech start-up companies in Germany, we have<br />
to realize that their future depends on basic research<br />
being as broad as possible and a great number of welltrained<br />
graduates and PhDs. Both goals can only be<br />
achieved with well-funded and highly flexible university<br />
institutions. It remains to be emphasized that basic<br />
biomedical research of the postgenome area in Germany<br />
will fall again behind the USA if the support<br />
of the institutions involved is not alike. The demand<br />
that basic biosciences at universities has to be given a<br />
higher priority than it receives at present has made it<br />
essential for the <strong>ZMBH</strong> to develop a mature dialogue<br />
with politicians and the public. We were very pleased<br />
that the Prime Minister of Baden-Wuerttemberg Erwin<br />
Teufel accepted our invitation to visit the <strong>ZMBH</strong> January<br />
on 21 st , 1999, on the occassion of his visit to the<br />
BioRegion and the Technology Park Heidelberg. Our<br />
State Minister for Economic Affairs Walter Doering<br />
visited the <strong>ZMBH</strong> on April 7 th , 1998 and discussed<br />
with us the aspects of know-how transfer and issues<br />
regarding start-up companies. Minister von Trotha discussed<br />
with us the above-mentioned issues during his<br />
visit of the <strong>ZMBH</strong> on July 23 rd , 1998. We discussed<br />
issues of the regulation regarding animal experimentation<br />
and gene technology with Members of the State<br />
Parliament Pfisterer and Hildebrand and Members of<br />
Hermann Bujard at the party of his 65 th birthday.<br />
19
transfers und Probleme von Grün<strong>der</strong>firmen zu diskutieren.<br />
Minister von Trotha diskutierte mit uns die<br />
oben erwähnten Themen während seines Besuches<br />
am 23.07.1998. Über aktuelle Fragen <strong>der</strong> gesetzlichen<br />
Regelungen von Tierversuchen und Gentechnik diskutierten<br />
wir mit den Mitglie<strong>der</strong>n des Landtags Hildebrand<br />
und Pfisterer und den Mitglie<strong>der</strong>n des Bundestags<br />
Binding und Dr. Lamers bei ihren Besuchen<br />
des <strong>ZMBH</strong>. Verschiedene Aspekte, die die Priorität<br />
<strong>der</strong> För<strong>der</strong>ung biowissenschaftlicher Grundlagenforschung<br />
betreffen, wurden auch beim Besuch des Mitglieds<br />
des Vorstandes <strong>der</strong> BASF <strong>für</strong> den Bereich Forschung<br />
Dr. Marcinowski am 19.11.198 behandelt.<br />
Der zweite Bereich <strong>der</strong> Kommunikation, um unserer<br />
Gesellschaft Themen <strong>der</strong> Biowissenschaften näher<br />
zu bringen, wurde mit Tagen <strong>der</strong> „Offenen Tür“,<br />
mit öffentlichen Vorträgen und Laborführungen sowie<br />
mit speziellen Veranstaltungen und Laborpraktika<br />
<strong>für</strong> Schüler angegangen. Wissenschaftler des <strong>ZMBH</strong><br />
Nobelpreisträger Bert Sakmann auf <strong>der</strong> Party zum 65.<br />
Geburtstag von Hermann Bujard.<br />
20<br />
waren auch an <strong>der</strong> Einrichtung <strong>der</strong> Ausstellung „Genwelten“<br />
im Landesmuseum <strong>für</strong> Technik und Arbeit in<br />
Mannheim, <strong>der</strong> Landesmesse „Wirtschaft trifft Wissenschaft“<br />
sowie <strong>der</strong> Landesinitiative „Zukunftswerkstatt<br />
Baden-Württemberg“ beteiligt.<br />
Das lebendige gesellschaftliche Leben im <strong>ZMBH</strong><br />
reichte wie<strong>der</strong>um vom Karneval bis zu Sommer- und<br />
Weihnachtsfeiern; nicht vergessen werden dürfen die<br />
von Axel Baumm wie immer sorgfältig geplanten<br />
jährlichen <strong>ZMBH</strong>-Ausflüge. Schwungvoll mit einer<br />
<strong>ZMBH</strong>-Party feierten wir auch den 65-jährigen<br />
Geburtstag von Hermann Bujard.<br />
Am 1. Januar 1998 endete die Amtszeit von Bernhard<br />
Dobberstein als Direktor des <strong>ZMBH</strong>. Als sein Nachfolger<br />
danke ich Bernhard Dobberstein, daß er dieses<br />
Amt zwei Jahre auf sich genommen und sich jetzt<br />
als zweiter stellvertreten<strong>der</strong> Direktor zur Verfügung<br />
gestellt hat. Ebenso bin ich Renato Paro dankbar, daß<br />
er bereits kurz nach seiner Berufung zustimmte, erster<br />
stellvertreten<strong>der</strong> Direktor zu werden und die vielen<br />
damit verbundenen Verwaltungsaufgaben zu übernehmen.<br />
Meinen Kollegen danke ich <strong>für</strong> ihre Unterstützung<br />
und das mir entgegengebrachte Vertrauen in den vergangenen<br />
32 Monaten meiner Direktorenschaft.<br />
Konrad Beyreuther<br />
the Bundestag Binding and Dr. Lamers during their<br />
visit of the <strong>ZMBH</strong>. Various aspects of the issue regarding<br />
the priority of basic biosciences at universities<br />
were also brought up during the visit of the Chief<br />
Scientific Officer of the BASF Dr. Marcinowski who<br />
came to the <strong>ZMBH</strong> on November 19 th , 1998. The<br />
second major area of communication to bring scientific<br />
issues to our community was addressed with<br />
“open days” with lectures and guided lab visits for<br />
the general public and with special meetings as well<br />
as lab courses for high school students. Scientists of<br />
the <strong>ZMBH</strong> were also actively involved in making the<br />
Exhibition “Gene World”, shown at the State Museum<br />
for Technology and Labour in Mannheim, a success.<br />
The social life at the <strong>ZMBH</strong> was again alive with<br />
annual highlights ranging from carneval to summer<br />
to Christmas parties; not to forget the annual <strong>ZMBH</strong><br />
excursion carefully organized by Axel Baumm. We<br />
also celebrated the 65 th birthday of Hermann Bujard<br />
with a <strong>ZMBH</strong> party.<br />
On January 1 st of 1998, Bernhard Dobberstein stepped<br />
down as director of the <strong>ZMBH</strong>. As his successor I<br />
thank Bernhard Dobberstein for shoul<strong>der</strong>ing this duty<br />
for two years and for carrying on as second deputy<br />
director. I am grateful also to Renato Paro for having<br />
accepted to become first deputy shortly after his<br />
appointment and for his commitment to take over the<br />
many administrative responsibilities.<br />
I thank my colleagues for their support and trust over<br />
the past 32 months of my directorship.<br />
Konrad Beyreuther<br />
21
Research reports<br />
23
Konrad Beyreuther<br />
Molecular Neurobiology of Alzheimer's<br />
Disease<br />
We study key aspects of Alzheimer’s disease related to<br />
synaptic and neuronal function, genetic and biochemical<br />
control mechanisms and how these are regulated<br />
in neural cells by the amyloid precursor protein (APP)<br />
supergene family, the presenilin (PS) gene family and<br />
other genes associated with neuronal function and the<br />
disease. The information that we gained is used to<br />
create cellular and animal models to study further the<br />
physiological and pathogenic role of the APP supergene<br />
family, and genes associated with cholesterol<br />
biosynthesis, transport, transmembrane signaling and<br />
metabolism in the brain.<br />
To un<strong>der</strong>stand synaptic loss and neurodegeneration in<br />
Alzheimer’s disease we have tried to consi<strong>der</strong> what<br />
are the physiological functions of the amyloid precursor<br />
protein (APP), its Aß-amyloid domain and of free<br />
Aß peptide. The latter is a normal metabolic product<br />
of APP and the principle subunit of amyloid plaques<br />
that are characteristic of Alzheimer‘s disease.<br />
From studies in transgenic Drosophila melanogaster<br />
and primary neurons, we suggest that in neurons<br />
APP’s physiological function is related to the regulation<br />
of synaptic strength whereas in nonneuronal<br />
cells APP appears to regulate cell-cell and cell-matrix<br />
adhesion.<br />
Since the axonal transport of APP is dependent on its<br />
Aß domain, this suggests that the Aß sequence could<br />
function as axonal sorting signal of APP. It also indicates<br />
that the Aß region could bind to molecules that<br />
control the recruitment of APP into axonally transported<br />
vesicles.<br />
In neurons, metabolism of APP releasing the Aß<br />
peptide was found to occur at all sorting stations of<br />
APP such as at the ER/cisGolgi and TGN/endosomes<br />
that gives rise to intracellular Aß peptide as well as<br />
at the cell surface leading to secretory Aß peptide.<br />
Regarding the Aß species generated in the different<br />
neuronal compartments, the long form of Aß (Aß42) is<br />
produced in the ER/cis Golgi and at or near to the cell<br />
surface and short Aß (Aß40) in the TGN/endosomal<br />
compartment and also at or near to the cell surface.<br />
Given an Aß function as axonal sorting signal of APP,<br />
release of Aß from APP may regulate the axonal transport<br />
of APP. Not only the removal of the Aß sequence<br />
from APP abolishes axonal APP transport but also free<br />
Aß could - by blocking the APP binding sites of the<br />
axonal transport machinery of APP - serve such a regulatory,<br />
physiological function. Excess intracellular<br />
and extracellular Aß may convert the latter physiological<br />
function of Aß to a pathogenic one by inhibiting<br />
the axonal transport of those proteins that use the<br />
same transport system as APP.<br />
Because the apoEε4 allele may be associated with<br />
higher cholesterol levels in neurons and higher risk<br />
of developing Alzheimer‘s disease and because the<br />
axonal transport of membrane proteins is cholesterol<br />
dependent, we studied the influence of cholesterol on<br />
neuronal Aß generation. By lowering the cholesterol<br />
level in neuronal cultures with statins (HMG-CoA<br />
reductase inhibitors), the formation of secretory and<br />
intracellular Aß is drastically reduced. Since the<br />
amount of Aß produced by neurons is cholesterol<br />
dependent, both the physiological and pathogenic regulation<br />
of APP transport by Aß appears to be controled<br />
in neurons by cholesterol, this implies a link<br />
between brain cholesterol, APP transport, Aß production<br />
and the risk of developing Alzheimer‘s disease.<br />
These intriguing relationships open new strategies to<br />
25
influence the progression of Alzheimer‘s disease by<br />
modulating cholesterol biosynthesis of neurons with<br />
statins.<br />
I. APP supergene family<br />
S. Eggert, S. Kreger, K. Paliga, A. Weidemann<br />
The Alzheimer‘s disease amyloid protein precursor<br />
(APP) gene is part of a multi-gene super-family from<br />
which sixteen homologous amyloidprecursor-like proteins<br />
(APLP) and APP species homologues are known.<br />
Comparison of exon structure (including the uncharacterised<br />
APL-1 gene), construction of phylogenetic<br />
trees, and analysis of the protein sequence alignment<br />
of known homologues of the APP super-family were<br />
performed to reconstruct the evolution of the family<br />
and to assess the functional significance of conserved<br />
protein sequences between homologues. This analysis<br />
supports an adhesion function for all members<br />
of the APP super family, with specificity determined<br />
by those sequences which are not conserved between<br />
APLP lineages, and provides evidence for an increasingly<br />
complex APP superfamily during evolution. The<br />
analysis also suggests that Drosophila APPL and Caenorhabditis<br />
elegans APL-1 may be a fourth APLP lineage<br />
indicating that these proteins, while not functional<br />
homologues of human APP, are similarly likely to regulate<br />
cell adhesion. Furthermore, the Aß sequence is<br />
highly conserved only in APP orthologues, strongly<br />
suggesting this sequence is of significant functional<br />
importance in this lineage.<br />
II. Physiological function of the Aß domain<br />
of APP and sites of production of Aß40<br />
and Aß42<br />
C. Bergmann, T. Hartmann, H. Grimm, P. J.<br />
Tienari, I. Tomic<br />
26<br />
Cleavage of APP by ß- and γ-secretase results in the<br />
release of the Aß domain. Cleavage occurs after residue<br />
40 of Aß (Aß40) and after residue 42 (Aß42).<br />
It is believed that even slightly increased amounts of<br />
Aß42 might be sufficient to cause Alzheimer‘s disease.<br />
What is the role of APP cleavage by these secretases.<br />
To un<strong>der</strong>stand the physiological function of APP<br />
processing centered around its Aß domain, deletion<br />
analyses were preformed which showed that the Aß<br />
domain of APP is essential for the axonal transport of<br />
APP (Fig. 1). This suggests that the Aß region of APP<br />
interacts with a sorting receptor or a sorting platform<br />
for delivery to the axonal membrane. Removal of the<br />
Aß domain or free Aß peptide is therefore expected<br />
to regulate the axonal transport of APP and other proteins<br />
utilizing the same transport machine as APP.<br />
Using immunogold electron microscopy and cell fractionation<br />
we have identified in neurons the endoplasmic<br />
reticulum/cis Golgi as the site for generation of<br />
Aß 42 and the trans-Golgi network (TGN) as the site<br />
for Aß 42 generation. It is interesting that intracellular<br />
generation of Aß seemed to be high in neurons,<br />
because we found that nonneuronal cells produced<br />
significant amounts of Aß40 and Aß42 only at the cell<br />
surface. This shows that neurons which are able to<br />
decode axonal sorting signals produce Aß at all sorting<br />
stations of APP. Aß has thus the potential to control<br />
neuronal APP transport. The specific production<br />
of the critical Aß42 isoform in the ER/cis Golgi of<br />
neurons links this compartment with APP transport<br />
and the generation of Aß. It also explains why primarily<br />
ER/cis Golgi localized (mutant) proteins such as<br />
the presenilins could induce Alzheimer’s disease. We<br />
suggest that the earliest event taking place in Alzheimer’s<br />
disease might be the generation of Aß42 in the<br />
ER.<br />
Figure 1: Aß is produced within neurons at the ER/cisGolgi as Aß42, at the TGN/endosomal compartment as Aß40 and at the<br />
cell surface/synapse as secreted Aß40 and Aß42. Secretory forms of Aß42 are aggregating to amyloid plaques. The Aß domain<br />
of APP is essential for axonal sorting of APP. Deletion of the extracellular part of Aß leads to somato-dendritic sorting and<br />
abolishes axonal sorting. Familial mutations at sites designated as FAD Swedish and FAD London increase Aß42 production<br />
but do not alter axonal sorting of APP.<br />
III. Mechanism of the cleavage of APP within<br />
its transmembrane domain by γ-secretase<br />
H. Grimm, B. Grziwa,T. Hartmann, S. Lichtenthaler<br />
Proteolytic processing of the amyloid precursor protein<br />
by ß-secretase yields A4CT (C99), which is<br />
cleaved further by the as yet unknown γ-secretase,<br />
yielding Aß40 and Aß42. We therefore used A4CT<br />
as a model to study the specificity of the cleavage of<br />
APP within its transmembrane domain by γ-secretase.<br />
Because the position of γ-secretase cleavage is cru-<br />
cial for the pathogenesis of Alzheimer‘s disease, we<br />
individually replaced all membrane-domain residues<br />
of A4CT outside the Aß domain with phenylalanine,<br />
stably transfected the constructs in COS7 cells, and<br />
determined the effect of these mutations on the<br />
cleavage specificity of γ-secretase (Aß42/Aß40 ratio).<br />
Assuming an alpha-helical conformation of the transmembrane<br />
domain of APP, mutations of residues<br />
superimposed at one helical face (residues 44, 47, and<br />
50) led to decreased Aß42/Aß40 ratios, whereas mutations<br />
affecting superimposed residues at the opposite<br />
site (residues 43, 45, 46, 49, and 51) led to increased<br />
27
Aß42/Aß40 ratios (Figure 2). A massive effect was<br />
observed for A4CT-I45F (34-fold increase) making<br />
this construct important for the generation of animal<br />
models for Alzheimer‘s disease. Unlike the other mutations,<br />
A4CT-V44F was processed mainly to Aß38, as<br />
determined by mass spectrometry. Our data provide a<br />
detailed model for the active site of γ-secretase (Figure<br />
2). According to this model Aß40 is produced when<br />
γ-secretase interacts with APP by binding to one side<br />
of the alpha-helical transmembrane domain of APP.<br />
Alternatively, Aß42 arises by binding of γ-secretase<br />
to the opposite side (Fig. 2). Mutations in the transmembrane<br />
domain of APP interfere with the interaction<br />
between γ-secretase and APP and, thus, alter the<br />
cleavage specificity of γ-secretase (Fig. 2).<br />
Figure 2: Schematic representation of the amino acid positions<br />
(P) of the ß-secretase product A4CT of APP relative to<br />
the cleavage site of γ-secretase. Top: linear arrangement of the<br />
residues relative to the cleavage site after residue 40 (Aß40)<br />
and residue 42 (Aß42). The scissile peptide bond is constituted<br />
by residues P1 and P1‘. Bottom: helical wheel arrangement<br />
of amino acids 40 to 49 of A4CT with respect to the cleavage<br />
sites after residues 40 and 42. Binding of γ-secretase to the<br />
transmembrane domain giving rise to Aß40 or Aß42 has to<br />
occur at opposite sides of the helix.<br />
28<br />
IV. Regulation of exon 15 splicing of APP<br />
premRNA<br />
C. Bergsdorff, S. Kreger, K. Paliga<br />
Alternative splicing of exon 15 of the amyloid precursor<br />
protein (APP) pre-mRNA generates two APP<br />
isoform groups APP(ex15) (containing exon 15) and<br />
L-APP (without exon 15), which show a cell-specific<br />
distribution in non-neuronal cells and neurons of rat.<br />
Both APP isoforms differ in regard to functional properties<br />
like post-translational modification, APP secretion,<br />
and proteolytic production of Aß peptide from<br />
APP molecules. Since Aß generation is an important<br />
factor in the development of Alzheimer‘s disease, one<br />
could anticipate that these major APP isoforms might<br />
contribute differentially to the mechanisms un<strong>der</strong>lying<br />
neurodegeneration in Alzheimer‘s disease. We<br />
established an APP minigene system in a murine cell<br />
system to identify cis-acting elements controlling exon<br />
15 recognition. A 12.5-kilobase pair genomic fragment<br />
of the murine APP gene contained all cis-regulatory<br />
elements to reproduce the splicing pattern of the<br />
endogenous APP transcripts. By using this approach,<br />
two intronic cis-elements flanking exon 15 were identified<br />
that block the inclusion of exon 15 in APP transcripts<br />
of non-neuronal cells. Point mutation analysis<br />
of these intronic regions indicated that pyrimidinerich<br />
sequences are involved in the splice repressor<br />
function. Finally, grafting experiments demonstrated<br />
that these regulatory regions cell-specifically enhance<br />
the blockage of a chimeric exon in the non-neuronal<br />
splicing system.<br />
V. Physiological function of APP<br />
A. Fossgreen, F. Reinhard, S. Scheurermann, P.<br />
Soba, J. Stumm<br />
In collaboration with B. Brückner and R. Paro, <strong>ZMBH</strong>,<br />
we used Drosophila melanogaster as a model system<br />
to analyze the function of APP by expressing wildtype<br />
and various mutant forms of human APP in fly<br />
tissue culture cells as well as in transgenic fly lines.<br />
After expression of full-length APP forms, secretion<br />
of APP but not of Aß was observed in both systems. By<br />
using SPA4CT, a short APP form in which the signal<br />
peptide was fused directly to the Aß region, transmembrane<br />
domain, and cytoplasmic tail, we observed<br />
Aß release in flies and fly-tissue culture cells. Consequently,<br />
we showed a γ-secretase activity in flies.<br />
Interestingly, transgenic flies expressing full-length<br />
forms of APP have a blistered-wing phenotype. As<br />
the wing is composed of interacting dorsal and ventral<br />
epithelial cell layers, this phenotype suggests that<br />
human APP expression interferes with cell adhesion/<br />
signaling pathways in Drosophila, independently of<br />
Aß generation. Wing morphogenesis in Drosophila is<br />
characterized by the apposition of two epithelial cell<br />
layers, the dorsal and ventral layer and may thus be<br />
used as a model to analyze the role of APP in the interaction<br />
of presynaptic and postsynaptic membranes at<br />
the synapse. We presume that APP negatively interacts<br />
with factors involved in the adhesion of the two wing<br />
epithelia and the two synaptic membranes, respectively.<br />
In Drosophila, some mutations deficient for the expression<br />
of specific integrin subunits develop distinct wing<br />
blisters whereas another one was shown to be involved<br />
in short term memory. This provides an interesting<br />
link between physiological APP functions, integrinmediated<br />
adhesion processes and memory mechanisms.<br />
Since integrins are heterodimeric receptors that<br />
may be ligand-bound to extracellular matrix molecules<br />
and may have a signaling function, integrins are<br />
especially required for the function of the cell-matrixcell<br />
junction, where one surface of a cell such as a<br />
synaptic density adheres to the other. This function<br />
of integrins supports our suggestion of APP’s involvement<br />
in cell adhesion. We envisage that APP might<br />
act as an antagonist for integrins, both in the developing<br />
wing and at plasticity of the synapse. APP could<br />
bind, either directly or via other molecules to the same<br />
extracellular matrix sites as integrins. Indeed, in rat<br />
primary neurons APP and integrins were colocalized<br />
in structures that are reminiscent of synapses. However,<br />
APP might be involved in other pathways resulting<br />
in a deterioration of the adhesion or signaling<br />
pathways supported by the integrins. For example,<br />
expression of a mutant form of G protein involved in<br />
cAMP signaling or the blistered gene encoding a protein<br />
related to human serum response factors result<br />
in Drosophila a comparable wing. Taken together,<br />
Drosophila seems to be a valuable animal-model<br />
system for studying the physiological and pathogenic<br />
functions of human APP since it may be suited for<br />
unraveling APP transmembrane signaling mechanisms<br />
potentially related to Alzheimer‘s disease. By using<br />
this fly model, testing genetic interactions between<br />
APP-expressing lines producing the blistered phenotype<br />
and other mutants engaged in wing morphology<br />
offers the perspective to unravel those physiological<br />
and pathogenic pathways linked to human APP.<br />
VI. Regulation of APP-Aß metabolism by cholesterol<br />
C. Bergmann, T. Hartmann, H. Runz, P. Semenfeld,<br />
I. Tomic<br />
Cholesterol metabolism and Alzheimer‘s disease are<br />
genetically linked. The apoEε4 allele of the apolipoprotein<br />
E gene is associated with higher cholesterol<br />
levels and was shown to increase the risk of develop-<br />
29
ing the disease. To study the influence of cholesterol<br />
on APP metabolism we collaborated with M. Simons,<br />
K. Simons and C.G. Dotti (EMBL) and used primary<br />
cultures of rat hippocampal neurons infected with<br />
recombinant Semliki Forest virus carrying APP and<br />
APP mutants. This system has been used previously<br />
by us to study the intracellular transport and processing<br />
of human APP. Inhibition of cholesterol biosynthesis<br />
and extraction of cholesterol from neuronal membranes<br />
was achieved by a combination of statin treatment<br />
and methyl-ß-cyclodextrin extraction. Statins<br />
such as lovastatin or simvastatin allow in the presence<br />
of low amounts of mevalonate the control of the synthesis<br />
of cholesterol by inhibiting 3-hydroxy-3-methylglutaryl-CoA<br />
(HMG-CoA) reductase. Mevalonate is<br />
required for the synthesis of non-steroidal products<br />
and essential to prevent inhibition of the proteasome<br />
by lovastatin. ß-Cyclodextrin has been shown to very<br />
efficiently and selectively extract cholesterol from the<br />
plasma membrane.<br />
The combined treatment of rat hippocampal neurons<br />
with statins and methyl-ß-cyclodextrin resulted in<br />
reduced intracellular Aß production and Aß secretion<br />
(Fig. 3), however APP levels and cell viability<br />
remained unaffected in both treated and control cells<br />
during the time course of the experiments.<br />
Since Aß generation occurs in two steps, we employed<br />
the two APP constructs APP695 and A4CT (SPC99)<br />
to analyze which Aß cleavage is inhibited by cholesterol<br />
lowering drugs. The first cleavage of APP by<br />
ß-secretase generates the 10-kDa fragment C99 that is<br />
further cleaved within the transmembrane domain by<br />
γ-secretase to produce Aß. When antibodies recognizing<br />
the C-terminal domain of APP were used to immunoprecipitate<br />
APP fragments from the cell lysates of<br />
cholesterol-depleted and control rat hippocampal neurons<br />
expressing human APP695 a dramatic inhibition<br />
30<br />
of ß-cleavage was detected while the production of the<br />
fragment generated by α-secretase was unperturbed.<br />
To analyze the effect of lovastatin treatment on the<br />
cleavage of γ-secretase, neurons expressing SPC99<br />
were used since removal of the transient signal<br />
sequence of SPC99 generates C99 which is identical<br />
with the C-terminal ß-secretase product of APP. Conversion<br />
of C99 to Aß requires γ-secretase. Both lovastatin<br />
treatment and methyl-ß-cyclodextrin treatment<br />
- alone and together - of neurons expressing SPC99<br />
inhibited Aß production as well suggesting that cholesterol-lowering<br />
regiments inhibit also γ-secretase<br />
cleavage. Thus our combined results suggest that cholesterol<br />
is required for APP cleavage by ß-secretase<br />
and γ-secretase but not for α-cleavage that produces<br />
αAPPsec. We conclude that lowering of cholesterol<br />
affects amyloidogenic processing of APP while allowing<br />
nonamyloidogenic cleavage to proceed.<br />
These findings raise the question how cholesterol<br />
affects Aß formation. One possibility is that reduc-<br />
Figure 3: Cholesterol-lowering of hippocampal neurons<br />
reduces intracellular (lysate) production and secretion<br />
(media) of Aß40 and Aß2. Neurons were grown for 4 days<br />
in the presence of lovastatin/mevalonate and after infection<br />
with Semliki Forest virus/APP-expression vectors were<br />
treated with 5mM methyl-ß-cyclodextrin for 20 min.<br />
tion in membrane cholesterol changes the intracellular<br />
transport of APP so that the protein does not reach the<br />
cellular sites where ß- and γ-secretase cleavage takes<br />
place. If this is the case, cholesterol is required as a<br />
sorting platform for inclusion of APP protein cargo<br />
destined for delivery to the apical membrane in nonneuronal<br />
cells and to the axonal membrane in neurons.<br />
Alternatively, the ß- and γ-secretase require cholesterol<br />
for their activity.<br />
VII. Aß as biological marker of Alzheimer’s<br />
disease<br />
A. Diehlmann, T. Hartmann, N. Ida<br />
All mutations known to cause familial Alzheimer‘s<br />
disease act by increasing the levels of soluble ß-amyloid<br />
peptide, especially the longer form, Aß42. However,<br />
in vivo elevation of soluble Aß in sporadic<br />
Alzheimer‘s disease has so far not been shown. In collaboration<br />
with M. Jensen and L. Lannfelt (Karolinska),<br />
we used our monoclonal antibodies specific for<br />
Aß40 and Aß42 in an enzyme-linked immunosorbent<br />
assay to investigate cerebrospinal fluid from sporadic<br />
Alzheimer‘s disease at different stages of disease<br />
severity, to clarify the roles of Aß42 and Aß40 during<br />
disease progression. We also evaluated three other<br />
groups, one group of patients with mild cognitive<br />
impairment who were at risk of developing dementia,<br />
a cognitively intact, nondemented reference group<br />
diagnosed with depression, and a perfectly healthy<br />
control group. We found that Aß42 is strongly elevated<br />
in early and mid stages of AD, and thereafter<br />
it declines with disease progression. On the contrary,<br />
Aß40 levels were decreased in early and mid stages<br />
of AD. The group of cognitively impaired patients<br />
and the depression reference group had significantly<br />
higher levels of Aß42 than the healthy control group,<br />
implying that Aß42 is increased not only in AD, but<br />
in other central nervous system conditions as well.<br />
Our data also point out the importance of having thoroughly<br />
examined control material. The initial increase<br />
and subsequent decrease of Aß42 adds a new biochemical<br />
tool to follow the progression of AD and<br />
might be important in the monitoring of therapeutics.<br />
Acknowledgements: We acknowledge the stimulating<br />
and productive collaboration with among others<br />
the group of Gerd Multhaup (<strong>ZMBH</strong>), Colin L. Masters<br />
(The University of Melbourne, Australia), Renato<br />
Paro (<strong>ZMBH</strong>), Kai Simons and Carlos G. Dotti<br />
(EMBL), Thomas A. Bayer (Psychiatry, University of<br />
Bonn, Germany), Michael Hennerici and Klaus Fassben<strong>der</strong><br />
(Neurology, University Heidelberg at Mann–<br />
heim), Lars Lannfelt (Karolinska Institute, Stockholm,<br />
Sweden); Joachim Schrö<strong>der</strong> (Psychiatry, University<br />
Heidelberg), Bart De Strooper (University Leuven,<br />
Belgium), Rudolph E. Tanzi and Ashley I. Bush<br />
(Harvard University, Cambridge, USA), Hans Peter<br />
Schmidt (Neuropathology, University Heidelberg),<br />
Hans Förstl (Psychiatry, TU Munich, Germany) and<br />
Christian Haass (Biochemistry, University Munich).<br />
External Funding<br />
Our research summarized in this report would not<br />
have been possible without the following grants: an<br />
institutional grant of the Minister for Science and Arts<br />
of the State of Baden-Württemberg, by grants and project<br />
grants of the Deutsche Forschungsgemeinschaft,<br />
of the Landes Forschungsschwerpunktprogramm of<br />
Baden-Württemberg, the European Community, of the<br />
German-Israelic-Foundation, of the Humboldt-Foundation,<br />
of the Fonds of the Chemical Industry of Germany<br />
and by generous donations.<br />
31
PUBLICATIONS<br />
Bayer, T., Cappai, R., Masters, C.L., Beyreuther, K.,<br />
and Multhaup, G. (1999). It all sticks together - the<br />
APP-related family of proteins and Alzheimer‘s disease.<br />
Molecular Psychiatry 4, 524-528.<br />
Bayer, T.A., Jäkälä, P., Hartmann, T., Egensperger,<br />
R., Buslei, R., Falkai, P., and Beyreuther, K. (1999).<br />
Neural expression profile of alpha-synuclein in developing<br />
human cortex. Neuroreport 10, 2799-2803.<br />
Bayer, T.A., Jäkälä, P., Hartmann, T., Havas, L.,<br />
McLean, C., Culvenor, J.G., Li, Q.X., Masters, C.L.,<br />
Falkai, P., and Beyreuther, K. (1999). Alpha-synuclein<br />
accumulates in Lewy bodies in Parkinson’s disease<br />
and dementia with Lewy bodies but not in Alzheimer’s<br />
disease beta-amyloid plaque cores. Neurosci.<br />
Lett. 266, 213-216.<br />
Beher, D., Elle, C., Un<strong>der</strong>wood, J., Davis, J.B., Ward,<br />
R., Karran, E., Masters, C.L., Beyreuther, K., and<br />
Multhaup, G. (1999). Proteolytic fragments of Alzheimer’s<br />
disease-associated presenilin 1 are present in<br />
synaptic organelles and growth cone membranes of rat<br />
brain. J. Neurochem. 72, 1564-1573.<br />
Bergsdorf, C., Paliga, K., Kreger, S., Masters, C.L.<br />
and Beyreuther, K. (<strong>2000</strong>). Identification of cis-Elements<br />
Regulating Exon 15 Splicing of the Amyloid<br />
Precursor Protein Pre-mRNA. J. Biol. Chem. 275,<br />
2046-2056.<br />
Borchert, T., Camakaris, J., Cappai, R., Masters, C.L.,<br />
Beyreuther, K., and Multhaup, G. (1999). Copper<br />
inhibits ß-amyloid production and stimulates the nonamyloidogenic<br />
pathway of amyloid-precursor-protein<br />
32<br />
secretion. Biochem. J. 344, 461-467.<br />
Capell, A., Grünberg, J., Pesold, B., Diehlmann, A.,<br />
Citron, M., Nixon, R., Beyreuther, K., Selkoe, D.J.,<br />
and Haass, C. (1998). The proteolytic fragments of<br />
the Alzheimer’s disease associated presenilin-2 form<br />
heterodimers and occur as a 100-150 kDa molecular<br />
weight complex. J. Biol. Chem. 273, 3205-3211.<br />
Cappai, R., Mok, S.S., Galatis, D., Tucker, D.F.,<br />
Henry, A., Beyreuther, K., Small, D.H., and Masters,<br />
C.L. (1999). Recombinant human amyloid precursor<br />
like-protein2 (APLP2) expressed in the yeast Pichia<br />
pastoris can stimulate neurite outgrowth. FEBS Lett.<br />
442, 95-98.<br />
Cappai, R., Stewart, L., Jobling, M.F., Thyer, J.M.,<br />
White, A.R., Beyreuther, K., Collins, S.J., Masters,<br />
C.L., Barrow, C.J. (1998). Familial prion disease<br />
mutation alters the secondary structure of recombinant<br />
mouse prion protein. Biochemistry 99, 3053-3058.<br />
Caswell, M.D., Mok, S.S., Henry, A., Cappai, R.,<br />
Klug, G., Beyreuther, K., Masters, C.L., and Small,<br />
D.H. (1999). The amyloid beta-protein precursor of<br />
Alzheimer’s disease is degraded extracellularly by a<br />
Kunitz protease inhibitor domain-sensitive trypsinlike<br />
serine protease in cultures of chick sympathetic<br />
neurons. Eur. J. Biochem. 266, 509-516.<br />
Cherny, R.A., Legg, J.T., McLean, C.A., Fairlie, D.P.,<br />
Huang, X., Atwood, C.S., Beyreuther, K., Tanzi,<br />
R.E., Masters, C.L., and Bush, A.I. (1999). Aqueous<br />
dissolution of Alzheimer’s disease Abeta amyloid<br />
deposits by biometal depletion. J. Biol. Chem. 274,<br />
23223-23228.<br />
Coulson, E.J., Paliga, K., Beyreuther, K., and Masters,<br />
C.L. (<strong>2000</strong>). What the evolution of the amyloid protein<br />
precursor supergene family tells us about its function.<br />
Neurochem. Int. 36, 175-184.<br />
Christie, G., Markwell, R.E., Gray, C.W., Smith, L.,<br />
Godfrey, F., Mansfield, F., Wadsworth, H., King, R.,<br />
McLaughlin, M., Cooper, D.G., Ward,R.V., Howlett,<br />
D.R., Hartmann, T., Lichtenthaler, F., Beyreuther, K.,<br />
Un<strong>der</strong>wood, J., Gribble, S.K., Cappai, R., Masters,<br />
C.L., Tamaoka, A., Gardner, R.L., Rivett, A.J., Karran,<br />
E.H., and Allsop, D. (1999). Alzheimer’s disease: correlation<br />
of the suppression of beta-amyloid peptide<br />
secretion from cultured cells with inhibition of the<br />
chymotrypsin-like activity of the proteasome. J. Neurochem.<br />
73, 195-204.<br />
Connop, B.P., Thies, R.L., Beyreuther, K., Ida, N.,<br />
and Reiner, P.B. (1999). Novel effects of FCCP [carbonyl<br />
cyanide p-(trifluoromethoxy)phenylhydrazone]<br />
on amyloid precursor protein processing. J. Neurochem.<br />
72, 1457-1465.<br />
Culvenor, J., Henry, A., Hartmann, T., Evin, G., Galatis,<br />
D., Friedhuber, A., Jaysena, U.L.H., Rajiv,<br />
Un<strong>der</strong>wood, J.R., Beyreuther, K., and Masters, C.L.<br />
(1998). Subcellular localization of the Alzheimer’s<br />
disease amyloid precursor protein and <strong>der</strong>ived peptides<br />
expressed in a recombinant yeast system. Amyloid 5,<br />
79-89.<br />
Culvenor, J.G., McLean, C.A., Cutt, S., Campbell,<br />
B.C., Maher, F., Jäkälä, P., Hartmann, T., Beyreuther,<br />
K., Masters, C.L., and Li, Q.X. (1999). Non-Abeta<br />
Component of Alzheimer’s Disease Amyloid (NAC)<br />
Revisited: NAC and alpha-Synuclein Are Not Associated<br />
with Abeta Amyloid. Am. J. Pathol. 155,<br />
1173-1181.<br />
Culvenor, J.G., Evin, G., Cooney, M.A., Wardan, H.,<br />
Sharples, R.A., Maher, F., Reed, G., Diehlmann, A.,<br />
Weidemann, A., Beyreuther, K., and Masters, C.L.<br />
(<strong>2000</strong>). Presenilin 2 expression in neuronal cells:<br />
induction during differentiation of embryonic carcinoma<br />
cells. Exp. Cell Res. 255, 192-206.<br />
Diehlmann, A., Ida, N., Weggen, S., Grunberg, J.,<br />
Haass, C., Masters, C.L., Bayer, T.A., and Beyreuther,<br />
K. (1999). Analysis of presenilin 1 and presenilin 2<br />
expression and processing by newly developed monoclonal<br />
antibodies. J. Neurosci. Res. 56, 405-419.<br />
Egensberger, R.,Weggen, S., Ida, N., Multhaup, G.,<br />
Schnabel, R., Beyreuther, K., and Bayer, T.A. (1999).<br />
Reverse relationsip between APP and ßA4 plaques in<br />
Down syndrome versus sporadic/familial Alzheimer’s<br />
disease. Acta Neuropathol. 97, 113-118.<br />
Emilien, G., Beyreuther, K., Masters, C.L., and Maloteaux,<br />
J.M. (<strong>2000</strong>). Prospects for pharmacological<br />
intervention in Alzheimer’s disease. Arch. Neurol. 57,<br />
454-459.<br />
Emilien, G., Maloteaux, J.M., Beyreuther, K., and<br />
Masters, C.L. (<strong>2000</strong>). Alzheimer’s disease: mouse<br />
models pave the way for therapeutic opportunities.<br />
Arch. Neurol. 57, 176-181.<br />
Evin, G., Reed, G., Tanner, J.A., Li, Y.-X., Culvenor,<br />
J.G., Fuller, S.J., Wadsworth, H., Allsop, D., Ward,<br />
R.V., Karran, E.H., Gray, C.W., Hartmann, T., Lichtenthaler,<br />
S., Weidemann, A., Beyreuther, K., and Masters,<br />
C.L. (1999). Characterization of gamma-secretase<br />
candidates from human brain using a new Western<br />
blot assay. In Alzheimer’s Disease and Related Disor<strong>der</strong>s.<br />
K. Iqbal, D.F. Swaab, B. Winblad, and H.M.<br />
33
Wisniewski eds. (John Wiley & Sons Ltd., Chichester),<br />
pp 441-417.<br />
Evin, G., Le Brocque, D., Culvenor, J.G., Galatis, D.,<br />
Weidemann, A., Beyreuther, K., Masters, C.L., and<br />
Cappai, R. (<strong>2000</strong>). Presenilin I expression in yeast<br />
lowers secretion of the amyloid precursor protein.<br />
Neuroreport 11, 405-408.<br />
Kwok, J.B., Li, Q.X., Hallupp, M., Whyte, S., Ames,<br />
D., Beyreuther, K., Masters, C.L., and Schofield, P.R.<br />
(<strong>2000</strong>). Novel Leu723Pro amyloid precursor protein<br />
mutation increases amyloid beta42(43) peptide levels<br />
and induces apoptosis. Ann. Neurol. 47, 249-253.<br />
Uhrig, R., Picard, M.A., Beyreuther, K., and Wiestler,<br />
M. (<strong>2000</strong>). Synthesis of antioxidative and anti-inflammatory<br />
drugs glucoconjugates. Carbohydr. Res. 325,<br />
72-80.<br />
Campbell, B.C., Li, Q.X., Culvenor, J.G. Jäkälä,<br />
P., Cappai, R., Beyreuther, K., Masters, C.L., and<br />
McLean, C.A. (<strong>2000</strong>). Accumulation of Insoluble<br />
alpha-Synuclein in Dementia with Levy Bodies. Neurobiol.<br />
Dis. 7, 192-<strong>2000</strong>.<br />
Fassben<strong>der</strong>, K., Masters, C.L., and Beyreuther, K.,<br />
(<strong>2000</strong>). Alzheimer‘s disease: An inflammatory disease?<br />
Neurobiol. Aging 21, 433-436.<br />
Jayasena, U.L., Gribble, S.K., McKenzie, A., Bey–<br />
reuther, K., and Masters, C.L. (<strong>2000</strong>). Identification of<br />
a unique conformational epitope in the carboxyl terminus<br />
of Alzheimer’s disease-associated ßA4 (1-42)<br />
amyloid using a monoclonal antibody. Ann. and Exp.<br />
Immunology (in press).<br />
34<br />
Ishii, K., Muelhauser, F., Liebl. U., Picard, M., Kuehl,<br />
S., Penke, B., Bayer, T., Hennerici, M., Beyreuther,<br />
K., Hartmann, T., and Fassben<strong>der</strong>, K. (<strong>2000</strong>). Subacute<br />
No generation induced by Alzheimer‘s ß-amyloid<br />
in the living brain: reversal by inhibition of the<br />
inducible NO synthase. FACEB J. 14, 1485-1489.<br />
Schönknecht, P., Schrö<strong>der</strong>, J., Pantel, J., Werle,<br />
E., Hartmann, T., Essig, M., Baudenstiel, K., and<br />
Beyreuther, K. (<strong>2000</strong>). Tauproteinspiegel in <strong>der</strong> Diagnostik<br />
<strong>der</strong> Alzheimer Demenz. Fortschr. Neurol. Psych.<br />
(in press).<br />
Fossgreen, A., Brückner, B., Czech, C., Paro, R.,<br />
Münch, G., Masters, C.L., and Beyreuther, K. (1998).<br />
Transgenic Drosophila expression of human Amyloid<br />
Precursor Protein (APP) in Drosophila melanogaster<br />
results in a blistered wing phenotype. Proc. Natl.<br />
Acad. Sci. USA 95, 13703-13708.<br />
Haß, S., Fresser, F., Köchl, S., Beyreuther, K., Utermann,<br />
G., and Baier, G. (1998). Physical interaction<br />
of ApoE with amyloid precursor protein independent<br />
of the amyloid Aß region in vitro. J. Biol. Chem. 273,<br />
13892-13897.<br />
Henry, A., Li, Q.X., Galatis, D., Hesse, L., Multhaup,<br />
G., Beyreuther, K., Masters, C.L., and Cappai, R.<br />
(1998). Inhibition of platelet activation by the Alzheimer’s<br />
diseases amyloid precursor protein. British Journal<br />
of Haematology 103, 402-414.<br />
Hock, C., Golombowski, S., Müller-Spahn, F., Naser,<br />
W., Beyreuther, K., Mönning, U., Schenk, D., Vigo-<br />
Pelfrey, C., Bush, A.M., Moir, R., Tanzi, R.E., Growdon,<br />
J., and Nitsch, R.M. (1998). Cerebrospinal fluid<br />
levels of amyloid precursor protein and amyloid<br />
ß-peptide in Alzheimer’s disease and major depression<br />
- Inverse correlation with dementia severity. Eur.<br />
Neurol. 39, 111-118.<br />
Jensen, M., Schrö<strong>der</strong>, J., Blomberg, M., Engvall, B.,<br />
Pantel, J., Ida, N., Basun, H., Wahlund, L.O., Werle,<br />
E., Jauss, M., Beyreuther, K., Lannfelt, L., and Hartmann,<br />
T. (1999). Cerebrospinal fluid A beta42 is<br />
increased early in sporadic Alzheimer’s disease and<br />
declines with disease progression. Ann. Neurol. 45,<br />
504-511.<br />
Jobling, M.F., Stewart, L.R., White, A.R., McLean,<br />
C., Friedhuber, A., Maher, F., Beyreuther, K., Masters,<br />
C.L., Barrow, C.J., Collins, S.J., and Cappai, R.<br />
(1999). The hydrophobic core sequence modulates the<br />
neurotoxic and secondary structure properties of the<br />
prion peptide 106-126. J. Neurochem. 4, 1557-1565.<br />
Multhaup, G., Hesse, L., Borchardt, T., Ruppert, T.,<br />
Cappai, R., Masters, C.L., and Beyreuther, K. (1998).<br />
Autoxidation of amyloid precursor protein and formation<br />
of reactive oxygen species. In Copper Transport<br />
and Its Disor<strong>der</strong>s: Molecular and Cellular Aspects. A.<br />
Leone and J.F.B Mercer eds. (New York: Plenum), pp<br />
75-94.<br />
LeBrocque, D., Henry, A., Cappai, R., Li, Q.X.,<br />
Tanner, J.E., Galatis, D., Gray, C., Holmes, S., Un<strong>der</strong>wood,<br />
J.R., Beyreuther, K., Masters, C.L., and Evin,<br />
G. (1998). Processing of the Alzheimer’s disease amyloid<br />
precursor protein in Pichia pastoris: α-, β- and<br />
γ-secretase products. Biochemistry 37, 14958-14965.<br />
Li, Q.-X., Cappai, R., Evin, G., Tanner, J.E., Gray,<br />
C.W., Beyreuther, K., and Masters, C.L. (1998). Products<br />
of the amyloid precursor protein generated by<br />
ß-secretase are present in human platelets, and secreted<br />
upon degranulation. American Journal of Alzheimer’s<br />
Disease. 15, 236-244.<br />
Li, Q.-X., Fuller, S.J., Beyreuther, K., and Masters, C.L.<br />
(1999). The amyloid precursor protein of Alzheimer’s<br />
disease in human brain and blood. J. Leukoc. Biol. 66,<br />
567-574.<br />
Li, Q.X., Maynard, C., Cappai, R., McLean, C.A.,<br />
Cherny, R.A., Lynch, T., Culvenor, J.G., Trevaskis,<br />
J., Tanner, J.E., Bailey, K.A., Czech, C., Bush, A.I.,<br />
Beyreuther, K., and Masters, C.L. (1999). Intracellular<br />
accumulation of detergent-soluble amyloidogenic<br />
A beta fragment of Alzheimer’s disease precursor protein<br />
in the hippocampus of aged transgenic mice. J.<br />
Neurochem. 72, 2479-2487.<br />
Li, Q.-X., Whyte, S., Tanner, J.E., Evin, G., Beyreuther,<br />
K., and Masters, C.L. (1998). Release of the amyloidogenic<br />
Aß fragment of Alzheimer’s disease from<br />
human platelets. Lab. Invest. 78, 461-469.<br />
Lichtenthaler, S.F., Multhaup, G., Masters, C.L., and<br />
Beyreuther, K. (1999). A novel substrate for analyzing<br />
Alzheimer’s disease gamma-secretase. FEBS Lett.<br />
453, 288-292.<br />
Lichtenthaler, S.F., Wang, R., Masters, C.L., and<br />
Beyreuther, K. (1999). Mechanism of the cleavage<br />
specificity of Alzheimer’s disease γ-secretase revealed<br />
by phenylalanine-scanning mutagenesis of the transmembrane<br />
domain of APP. Proc. Natl. Acad. Sci. USA<br />
96, 3053-3058.<br />
Masters, C.L., and Beyreuther, K. (1998), Science,<br />
medicine and the future. Alzheimer’s disease. British<br />
35
Medical Journal 316, 446-448.<br />
Masters, C.L., and Beyreuther, K. (1999). Amyloid<br />
Nomenclature Committee. Amyloid 6, 151-152.<br />
Masters, C.L., and Beyreuther, K. (<strong>2000</strong>). Genomic<br />
neurology. Arch. Neurol. 57, 53.<br />
McLean, C.A., Cherry, R.A., Fraser, F.W., Fuller, S.J.,<br />
Smith, M.J., Beyreuther, K., Bush, A.I., and Masters,<br />
C.L. (1999). Soluble pool of Aß as a determinant of<br />
severity of neurodegeneration in Alzheimer’s disease.<br />
New Engl. J. Medicine, Ann. Neurol. 46, 860-866.<br />
Moir, R.D., Lynch, T., Bush, A.I., Whyte, S., Henry,<br />
A., Portbury, S., Tanzi, R.E., Multhaup, G., Small,<br />
D.H., Beyreuther, K., and Masters, C.L. (1998). Relative<br />
increase in Kunitz protease inhibitory forms of<br />
cerebral Aß amyloid protein precursor in Alzheimer’s<br />
disease. J. Biol. Chem. 273, 5013-5019.<br />
Multhaup, G., Hesse, L., Borchardt, T., Ruppert, T.,<br />
Cappai, R., Masters, C.L., Beyreuther, K., and Masters,<br />
C.L. (1999). Autoxidation of amyloid precursor<br />
protein and formation of reactive oxygen species. Adv.<br />
Exp. Med. Biol. 448, 183-192.<br />
Multhaup, G., Masters, C.L., and Beyreuther, K.<br />
(1998). Oxidative stress in Alzheimer’s disease. Alzheimer’s<br />
Reports 1, 147-154.<br />
Multhaup, G., Masters, C.L., Beyreuther, K., and<br />
Cappai, R. (1998). The biological activities and function<br />
of the amyloid precursor protein of Alzheimer’s<br />
disease. In The Molecular Biology of Alzheimer’s Disease<br />
- Genes and Mechanisms Involved in Amyloid<br />
Generation. C. Hasss, ed. (Harwood Academic Publishers,<br />
Amsterdam), pp 75-94.<br />
36<br />
Multhaup, G., Ruppert, T., Schlicksupp, A., Hesse, L.,<br />
Bill, E., Pipkorn, R., Masters, C.L., and Beyreuther,<br />
K. (1998). Copper-binding amyloid precursor protein<br />
un<strong>der</strong>goes a site-specific fragmentation in the reduction<br />
of hydrogen peroxide. Biochem. 37, 7224-7230.<br />
Postuma, R.B., Martins, R.N., Cappai, R., Beyreuther,<br />
K., Masters, C.L., Strickland, D.K., Mok, S.S., and<br />
Small, D.H. (1998). Effects of the amyloid protein<br />
precursor of Alzheimer’s disease and other ligands of<br />
the LDL receptor-related protein on neurite outgrowth<br />
from sympathetic neurons in culture. FEBS Lett. 428,<br />
13-16.<br />
Postuma, R.B., He, W., Nunan, J., Beyreuther, K.,<br />
Masters, C.L., Barrow, C.J., and Small, D.H. (<strong>2000</strong>).<br />
Substrate-bound beta-amyloid peptides inhibit cell<br />
adhesion and neurite outgrowth in primary cultures. J.<br />
Neurochem. 74, 1122-1130.<br />
Ren, R.F., Lah, J.J., Diehlmann, A., Kim, E.S., Hawver,<br />
D.B., Levey, A.I., Beyreuther, K., and Flan<strong>der</strong>s, K.C.<br />
(1999). Differential effects of transforming growth<br />
factor-beta(s) and glial cell line-<strong>der</strong>ived neurotrophic<br />
factor on gene expression of presenilin-1 in human<br />
post-mitotic neurons and astrocytes. Neuroscience 93,<br />
1041-1049.<br />
Rondot, S., Anthony, K.G., Duebel, S., Ida, N., Wiemann<br />
S., Beyreuther, K., Frost, L.S., Little, M., and<br />
Breitling, F. (1998). Epitopes fused to F-pilin are<br />
incorporated into functional recombinant pili. J. Mol.<br />
Biol. 279, 589-603.<br />
Rossjohn, J., Cappai, R., Feil, S.C., Henry, A., McKinstry,<br />
W.J., Galatis, D., Hesse, L., Multhaup, G.,<br />
Beyreuther, K., Masters, C.L., and Parker, M.W.<br />
(1999). Crystal structure of the N-terminal, growth<br />
factor-like domain of Alzheimer amyloid precursor<br />
protein. Nat. Struct. Biol. 6, 327-331.<br />
Sberna, G., Sáez-Valero, Li, Q.-X., Czech, C.,<br />
Beyreuther, K., Masters, C.L., McLean, C.A., and<br />
Small, D.H. (1998). Acetylcholinesterase is increased<br />
in the brain of transgenic mice expressing the C-terminal<br />
fragment (CT100) of the ß-amyloid protein precursor<br />
of Alzheimer’s disease. J. Neurochemistry 71,<br />
723-731.<br />
Simons, M., Keller, P., De Strooper, B., Beyreuther,<br />
K., Dotti, C.G., and Simons, K. (1998). Chostersterol<br />
depletion inhibits the generation of ß-amyloid in hippocampal<br />
neurons. Proc. Natl. Acad. Sci. USA 95,<br />
6460-6464.<br />
Smith, M.J., Gardner, R.J., Knight, M.A., Forrest,<br />
S.M., Beyreuther, K., Storey, E., McLean, C.A.,<br />
Cotton, R.G., Cappal, R., and Masters, C.L. (1999).<br />
Early-onset Alzheimer’s disease caused by a novel<br />
mutation at codon 219 of the presenilin-1 gene. Neuroreport<br />
10, 503-507.<br />
Storey, E., Katz, M., Brickman, Y., Beyreuther, K.,<br />
and Masters, C.L. (1999). Amyloid precursor protein<br />
of Alzheimer’s disease: evidence for a stable, fulllength,<br />
trans-membrane pool in primary neuronal cultures.<br />
Eur. J. Neurosci. 11, 1779-1788.<br />
Tesco, G., Kim, T.-W., Diehlmann, A., Beyreuther,<br />
K., and Tanzi, R.E. (1998). Abrogation of the presenilin<br />
1/ß-catenin interaction and preservation of the<br />
heterodimeric presenilin 1 complex following caspase<br />
activation. J. Biol. Chem. 273, 33909-33914.<br />
Weggen, S., Diehlmann, A., Buslei, R., Beyreuther,<br />
K., and Bayer, T. (1998). Prominent Expression of<br />
Presenilin-1 in Senile Plaques and Reactive Astrocytes<br />
in Alzheimer’s Disease Brain. Neuroreport 9,<br />
3279-3283.<br />
Weggen, S., Tienari, P.J., Beyreuther, K., and Bayer<br />
T.A. (1999). Retroviral shuttle vectors in Alzheimer’s<br />
disease research. Neurol. Psych. Brain Res. 6,<br />
213-218.<br />
Weidemann, A., Paliga, K., Durrwang, U., Reinhard,<br />
F.B.M., Evin, G., Masters, C.L., and Beyreuther, K.<br />
(1999). Cleavage of the Alzheimer’s disease amyloid<br />
precursor protein during apoptosis by activated caspases.<br />
In Alzheimer’s Disease and Related Disor<strong>der</strong>s.<br />
K. Iqbal, D.F. Swaab, B. Winblad, and H.M. Wisniewski<br />
eds. (John Wiley & Sons Ltd., Chichester), pp<br />
391-396.<br />
White, A.R., Bush, A.I., Beyreuther, K., Masters, C.L.,<br />
and Cappai, R. (1999). Exacerbation of copper toxicity<br />
in primary neuronal cultures depleted of cellular<br />
glutathione. J. Neurochem. 72, 2092-2098.<br />
White, A.R., Collins, S.J., Maher, F., Jobling, M.F.,<br />
Stewart, L.R., Thyer, J.M., Beyreuther, K., Masters,<br />
C.L., and Cappai, R. (1999). Prion protein-deficient<br />
neurons reveal lower gluthione reductase activity and<br />
increased susceptibility to hydrogene peroxide toxicity.<br />
Am. J. Pathol. 155, 1723-1730.<br />
White, A.R., Multhaup, G., Maher, F., Bellingham,<br />
S., Camakaris, J., Zheng, H., Bush, A.I., Beyreuther,<br />
K., Masters, C.L., and Cappai, R. (1999). The Alzheimer’s<br />
disease amyloid precursor protein modulates<br />
copper-induced toxicity and oxidative stress in primary<br />
neuronal cultures. J. Neurosci. 19, 9170-9179.<br />
37
White, A.R., Reyes, R., Mercer. K-F-B-. Camacaris,<br />
J., Zheng, H., Bush, A.I., Multhaup, G., Beyreuther,<br />
K., Masters, C.L., and Cappai, R. (1999). Copper<br />
levels are increased in the cerebral cortex and liver<br />
of APP and APLP2 knockout mice. Brain Res. 842,<br />
439-444.<br />
White, A.R., Zheng, H., Galatis, D., Maher, F., Hesse,<br />
L., Multhaup, G., Beyreuther, K., Masters, C.L., and<br />
Cappai, R. (1998). Survival of cultured neurons from<br />
amyloid precursor protein knock-out mice against<br />
Alzheimer’s amyloid-ß toxicity and oxidative stress.<br />
J. Neurosci. 18, 6207-6217.<br />
THESES<br />
Diploma<br />
Großkreutz, Yannik (1999). Herstellung und Charakterisierung<br />
von APP-GFP Fusionsproteinen.<br />
Scheuermann, Stefan (1999). Nachweis und Charakterisierung<br />
<strong>der</strong> Dimerisierung des Amyloid Precursor<br />
Proteins.<br />
Dissertations<br />
Bergsdorf, Christian (1999). Molekularbiologische<br />
Charakterisierung <strong>der</strong> Regulation des alternativ gespleißten<br />
Exons 15 des Amyloid Vorläuferproteins.<br />
Picard, Martin (1998). Untersuchungen zur Alzheimer<br />
Krankheit: Neuroinflammation und Glucose Konjugate<br />
entzündungshemmen<strong>der</strong> Wirkstoffe.<br />
Uhrig, Rainer (1999). Glycokonjugate von Antioxidantien<br />
– Synthese, Transport und Bedeutung <strong>für</strong> ein<br />
hirnspezifisches Drug Targeting bei Morbus Alzheimer.<br />
38<br />
STRUCTURE OF THE GROUP<br />
E-mail: beyreuther@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Beyreuther, Konrad, Prof.<br />
Dr. rer. nat. Dr. med. h. c.<br />
Assistant Hartmann, Tobias, Dr. rer. nat.<br />
Senior research<br />
associates Prior, Peter, Dr. rer. nat.<br />
Weidemann, Andreas, Dr. rer. nat.<br />
Postdoctoral<br />
fellows Bergmann, Christine, Dr. med.<br />
Bergsdorf, Christian, Dr. rer. nat.<br />
Foßgreen, Anke, Dr. rer.nat.*<br />
Ishii, Kazuhiro, M.D.<br />
Lichtenthaler, Stefan, Dr. rer. nat.*<br />
Paliga, Krzysztof, Dr. rer. nat.<br />
Peraus, Gisela, Dr. rer.nat*<br />
Picard, Martin, Dr. rer. nat.*<br />
Uhrig, Rainer, Dr. rer. nat.*<br />
Zhao, Liyun, Ph.D.<br />
Visiting<br />
scientist Jäkkälä, Pekka, M.D.*<br />
Graduate<br />
students Grimm, Heike, Dipl. Biol.<br />
Grziwa, Beate, Dipl. Biol.<br />
Kinter, Jochen, Dipl. Biol.*<br />
Reinhard, Friedrich, Dipl. Chem.<br />
Runz, Heiko, cand. med.*<br />
Scheuermann, Stefan, Dipl. Biol.<br />
Soba, Peter, Dipl. Chem.*<br />
Stumm, Joachim, Dipl. Biol.<br />
Staff scientist Diehlmann, Anke, Dipl. Biol.*<br />
Techn.<br />
assistants Kreger, Sylvia<br />
Samenfeld, Petra<br />
Tomic, Inge<br />
Administration<br />
and secretary Demuth, Heidemarie<br />
*part of the time reported<br />
39
Project Group Gerd Multhaup<br />
Ligand-Dependent Functions of the Amyloid<br />
Precursor Protein (APP) and Ligand-Associated<br />
Conformational Changes<br />
APP is a transmembrane glycoprotein that un<strong>der</strong>goes<br />
extensive alternative splicing. APP belongs to a multigene<br />
family that contains at least two other homologs<br />
known as amyloid precursor-like proteins (APLP1 and<br />
APLP2) (Fig. 1). APP and APLPs share most of the<br />
domains and motifs of APP, but only APP contains the<br />
Aß region and can be cleaved by ß- and γ-secretase<br />
to generate Aß. Thus, APLPs cannot contribute to Aß<br />
deposition in Alzheimer’s disease but may compensate<br />
for the function of APP.<br />
The normal functions of APP and APLPs are not<br />
well un<strong>der</strong>stood. There exists at least some evidence<br />
for neuritic and protective roles. APP binds Zn(II) at<br />
higher nanomolar concentrations and an altered APP<br />
metabolism or expression level is believed to result in<br />
neurotoxic processes. APP can reduce Cu(II) to Cu(I)<br />
in a cell-free system potentially leading to increased<br />
oxidative stress in neurons. The domain that contributes<br />
to such activities is the copper binding domain<br />
residing between residues 135 and 158 of APP, a<br />
region that shows strong homology to APLP2 but not<br />
to APLP1.<br />
Potentially, APP-Cu(I) complexes are involved that<br />
reduce hydrogen peroxide to form an APP-Cu(II)hydroxyl<br />
radical intermediate. APP residues 135 to<br />
158 consisting of cysteine and Cu-coordinating histidine<br />
residues can modulate copper-mediated lipid<br />
peroxidation and neurotoxicity in culture of APP<br />
knockout (APP 0/0 ) and wild-type (wt) neurons. Wt<br />
neurons were found to be more susceptible than APP 0/0<br />
neurons to physiological concentrations of copper but<br />
not other metals.<br />
Figure 1: Summary of the structural domains and binding motifs of the APP-family. The broken vertical lines denote APP<br />
sequences conserved in the APLP molecules. Abbreviations used are: α, alpha-secretase site; “α”, homologous to alphasecretase<br />
cleavage site for release of the ectodomain; β, beta-secretase site; CD, cytoplasmic domain; CBD, collagen-binding<br />
domain; CHO, N-linked carbohydrate site; CS-GAG, chondroitin sulfate glycosaminglycan; CuBD-1, copper-binding domain<br />
1; CuBD-2, copper-binding domain 2; cys, cysteine; δ, delta-secretase site; γ, gamma-secretase site; GPD, growth promoting<br />
domain; HBD-1, heparin-binding domain 1; HBD-2, heparin-binding domain 2; KPI, Kunitz-type protease inhibitor; NPXY,<br />
asparagine-proline-any amino acid-tyrosine; OX-2, OX-2 homology domain; Poly-T, poly-threonine; P, phosphorylation site;<br />
SP, signal peptide; ZnBD-1, zinc-binding domain 1; ZnBD-2, zinc-binding domain 2.<br />
40<br />
APP 0/0 mice have significantly increased copper but<br />
not zinc or iron levels in the cerebral cortex and liver<br />
as compared to age and genetically matched wt mice.<br />
APLP2 0/0 mice also revealed increases in copper in<br />
cerebral cortex and liver. These findings suggest that<br />
the APP family can modulate copper homeostasis<br />
and that APP/APLP2 expression may be involved in<br />
copper efflux from liver and cerebral cortex. Most<br />
importantly, copper was found to influence APP processing<br />
in a cell culture model system when copper<br />
was observed to greatly reduce the levels of amyloid<br />
Aß peptide and copper also caused an increase in<br />
the secretion of the APP ectodomain. An increase in<br />
intracellular APP levels which paralleled the decrease<br />
in Aß generation suggested that additional copper was<br />
acting on two distinct regulating mechanisms, one on<br />
Aß production and the other on APP synthesis. Taken<br />
together, APP and APLP2 are most likely involved in<br />
copper homeostasis.<br />
Our work is funded by grants of the DFG (through<br />
SFB317, 1991-1999; Mu-901), by the BMBF, by the<br />
AFI-foundation (Alzheimer Forschung Initiative), by<br />
the Graduiertenkolleg (C. Elle), by the International<br />
Copper Association (ICA) and by the Fonds <strong>der</strong> Chemischen<br />
Industrie.<br />
We collaborate with Prof. Dr. Dr. K. Beyreuther<br />
(<strong>ZMBH</strong>, Heidelberg), Prof. Dr. C.L. Masters (University<br />
of Melbourne, Dep. of Pathology), Prof. Dr. C.<br />
Haass (Ludwig-Maximilians-University, Adolf-Butenandt-Institute,<br />
München), Prof. Dr. W. Stremmel (University<br />
of Heidelberg, Dep. of Gastroenterology), Prof.<br />
Dr. J. Camakaris (University of Melbourne, Dep. of<br />
Genetics), Prof. Dr. J. Mercer (Deakin University,<br />
Melbourne), Prof. Dr. A. Bush (Harvard University,<br />
USA), Dr. T. Bayer (University of Bonn) and Prof. Dr.<br />
K. Wieghardt (MPI, Mülheim).<br />
I. The role of copper in the pathological<br />
function of the amyloid precursor protein<br />
(APP)<br />
C. Schmidt, M. Strauss, A. Simons, A. Schlicksupp,<br />
G. Multhaup (in collaboration with T.<br />
Bayer, R. Cappai, A. White, E. Bill, R. Pipkorn,<br />
C.L. Masters and K. Beyreuther)<br />
The extracellular domain of transmembrane Aß amyloid<br />
precursor protein (APP) has a Cu(II) reducing<br />
activity upon Cu(II) binding associated with the formation<br />
of a new disulfide bridge. The complete assignment<br />
of the disulfide bond revealed the involvement<br />
of cysteines 144 and 158 around copper-binding histidine<br />
residues ( Fig. 2).<br />
Figure 2: The APP copper binding motif corresponds to<br />
type-II copper binding sites and is encompassed by copper<br />
coordinating histidine residues 147, 149 and 151. The<br />
reduction of copper by APP results in a corresponding oxidation<br />
of cysteines 144 and 158. The zinc binding motif<br />
maps to exon 5 of APP between residues 181-200 with the<br />
neighboring cysteines involved in chelation.<br />
41
APP catalyzed the reduction of H 2 O 2 and oxidation of<br />
Cu(I) to Cu(II) in a „peroxidative“ reaction in vitro.<br />
The resulting bound copper-hydroxyl radical intermediate<br />
[APP-Cu(II) (•OH)] then likely participated in<br />
a Fenton type of reaction with radical formation as a<br />
prerequisite for APP-Cu(I) complex degradation. Evidence<br />
from two observations suggests that the reaction<br />
takes place in two phases. Bathocuproine, a trapping<br />
agent for Cu(I), abolished the initial fragmentation<br />
of APP, and chelation of Cu(II) by DTPA (diethylenetriaminepentaacetic<br />
acid) interrupted the reaction<br />
cascade induced by H 2 O 2 at later stages. WT cortical,<br />
cerebellar and hippocampal neurons were significantly<br />
more susceptible than their respective APP 0/0 neurons<br />
to toxicity induced by physiological concentrations<br />
of copper but not by zinc or iron. There was no difference<br />
in copper toxicity between APLP2 0/0 and WT<br />
neurons, demonstrating specificity for APP-associated<br />
copper toxicity. Treatment of neuronal cultures with a<br />
peptide corresponding to the human APP copper binding<br />
domain (APP142-166) potentiated copper but not<br />
iron or zinc toxicity. Incubation of APP142-166 with<br />
low density lipoprotein (LDL) and copper resulted<br />
in significantly increased lipid peroxidation compared<br />
to copper and LDL alone. Substitution of the<br />
copper co-ordinating histidine residues with asparagines<br />
(APP142-166 H147N, H149N, H151N ) abrogated the<br />
toxic effects. A peptide corresponding to the zinc binding<br />
domain (APP181-208) failed to induce copper or<br />
zinc toxicity in neuronal cultures. These data support a<br />
role for the APP copper binding domain in APP-mediated<br />
Cu(I) generation and toxicity in primary neurons.<br />
To test the hypothesis that biometals such as copper<br />
can contribute to the amyloid pathology in AD we are<br />
currently investigating the dietary exposure of copper<br />
and its chelators to APP transgenic mice and to APP<br />
42<br />
knock-out mice. The effects of dietary supplementation<br />
is studied by ICP-MS (inductively coupled plasma<br />
mass-spectrometry) to determine the levels of Cu, Zn<br />
and Fe. The AD pathology is analyzed by conventional<br />
methods. We expect that Cu has an effect on the<br />
AD pathology and metal-chelation can delay amyloid<br />
Aß deposition in APP transgenic mice.<br />
II. The role of copper and zinc in the normal<br />
function of the amyloid precursor protein<br />
(APP)<br />
C. Schmidt, A. Simons, M. Strauss, A. Schlicksupp,<br />
G. Multhaup (in collaboration with D.<br />
Ha-Hao, R. Cappai, A. White, C.L. Masters and<br />
K. Beyreuther)<br />
The expression of APP and APLP2 in the brain<br />
suggests they could have an important direct or indirect<br />
role in neuronal metal homeostasis. In APP and<br />
APLP2 knockout mice copper levels were significantly<br />
elevated in both APP 0/0 and APLP2 0/0 cerebral<br />
cortex (40% and 16%, respectively) and liver (80% and<br />
36%, respectively) compared with matched wild-type<br />
(WT) mice. These findings indicate APP and APLP2<br />
expression specifically modulates copper homeostasis<br />
in the liver and cerebral cortex, the latter being a<br />
region of the brain particularly involved in AD. Perturbations<br />
to APP metabolism and in particular, its secretion<br />
or release from neurons may alter copper homeostasis<br />
and explain a disturbed metal-ion homeostasis<br />
observed in AD.<br />
Zinc up to concentrations of 50µM or the presence of<br />
1,10-phenanthroline specifically increased the level of<br />
secreted APP in APP transfected CHO-K1 cells. By<br />
contrast, the level of secreted APP in copper-resistant<br />
CHO-CUR3 cells remained unaffected. APP holoprotein<br />
increased dramatically in CHO-CUR3 cells com-<br />
pared with CHO-K1 cells. The large decrease of Aß<br />
release seen in both cell lines at elevated extracellular<br />
zinc levels was due to specific inhibition of secretion.<br />
These results indicate that a disturbed zinc-homeostasis<br />
may be an important factor influencing APP production,<br />
transport and processing.<br />
Figure 3: The α- and β-secretase protease activities cleave<br />
APP within its ectodomain. The remaining membranebound<br />
C-terminal fragment p3CT (after α-secretase cleavage<br />
of APP) is further cleaved by γ-secretase in the middle<br />
of the putative transmembrane domain, yielding p3 (α- and<br />
γ-secretase activities). Amyloid Aβ is produced by β- and<br />
γ-secretase activities; CuBD-I, copper binding domain 1.<br />
Adding copper to APP-transfected CHO cells greatly<br />
reduced the levels of ß-amyloid (Aß) peptide in both<br />
parental CHO-K1 and in copper resistant CHO-CUR3<br />
cells which have lower intracellular copper levels.<br />
Copper also caused an increase in the secretion of the<br />
APP ectodomain indicating that the large decrease in<br />
Aß release was not due to a general inhibition in protein<br />
secretion. There was an increase in intracellular<br />
full-length APP levels which paralleled the decrease<br />
in Aß generation suggesting the existence of two distinct<br />
regulating mechanisms, one acting on Aß production<br />
and the other on APP synthesis. Thus, our<br />
findings suggest that copper or copper agonists might<br />
be useful tools to discover novel targets for anti-<br />
Alzheimer drugs since copper promoted the non-amyloidogenic<br />
pathway of APP.<br />
Our current un<strong>der</strong>standing is that copper and/or zinc<br />
binding is central to the normal cellular function of<br />
APP. We therefore want to identify agonists of APP<br />
ligand binding sites (e.g. of the copper-binding site)<br />
that are able to inhibit amyloidogenic proteolytic processing<br />
of APP. Lead compounds will be discovered<br />
followed by a further screen for small APP ligands.<br />
Our research program combines biochemical, immunohistochemical<br />
and cellular approaches to start a<br />
risk-benefit analysis of divalent metal binding agonists<br />
of APP in vitro.<br />
III. Dimerization and stability of APP isoforms:<br />
influence of APP ligands on relative<br />
stability and metabolism<br />
M. Strauss, C. Schmidt, A. Simons, A. Schlicksupp,<br />
C. Elle, G. Multhaup (in collaboration with<br />
C. Haass, C.L. Masters and K. Beyreuther)<br />
The highly conserved nature and tissue specificity of<br />
the eight APP isoforms provide circumstantial evidence<br />
that functional differences among isoforms may<br />
exist in vitro and in vivo. When APP and Aß are central<br />
to AD pathogenesis, then molecules which influence<br />
the conformation and the stability of APP isoforms<br />
and that interact with specific APP forms could<br />
differentially alter their metabolism/activity and thus<br />
represent risk factors for AD.<br />
For example, heparin and Zn(II) were found to augment<br />
the ability of full-length and secreted KPI-APP to<br />
inhibit FXIa. In contrast, both compounds heparin and<br />
43
Zn(II) failed to have an effect on C-terminally truncated<br />
recombinant KPI-APP (APP residues 18-350)<br />
from Pichia pastoris and on the inhibition of trypsin<br />
or chymotrypsin. Together with the observation that<br />
native KPI-APP was required for the potentiation of<br />
inhibition by Zn(II) these data indicate that inhibitory<br />
effects were enhanced by other domains of APP than<br />
KPI. Further structural investigations of the N-terminal<br />
heparin binding domain of APP (residues 28-123)<br />
revealed a highly charged basic surface and an abutting<br />
hydrophobic surface immediately adjacent to the<br />
low-affinity N-terminal heparin binding site (residues<br />
96-110) that is proposed to play an important function<br />
in dimerization and/or ligand binding. A second heparin<br />
binding site of high affinity was identified within<br />
a region conserved in rodent and human APP, APLP1<br />
and APLP2. This binding site was located between<br />
residues 316-337 of APP695 which is the carbohydrate<br />
domain of APP (Figure 1). The affinity for heparin<br />
is increased two- to four-fold in the presence of<br />
micromolar Zn(II).<br />
The current model of APP dimerization is complex<br />
and the information about the nature of APP-APP<br />
interaction is indirect and inferred. Indeed, APP<br />
released into the cell culture supernatant has been<br />
found to be secreted as dimers and also to exist intracellularly<br />
as dimerized transmembrane APP. Most<br />
likely, both sites, the APP-APP interaction domain<br />
encoded by residues 448-465 and the large hydrophobic<br />
patch of N-terminal APP residues 96-116 (N-terminal<br />
interaction site, (NIS)) are involved in the generation<br />
of dimers (Fig. 4). The N-terminal site is sufficient<br />
for oligomerization since dynamic light scattering<br />
analysis of recombinant APP18-350 from Pichia<br />
pastoris showed that the C-terminal truncated protein<br />
is secreted as a homodimer (Fig. 4).<br />
44<br />
Figure 4: The three-dimensional structure of an N-terminal<br />
fragment (APP residues 28-123) has been determined<br />
(Rossjohn et al., 1999) and resembles a growth factor. The<br />
large hydrophobic patch of residues 96-116 is most likely<br />
involved in the generation of dimers (arrows). The structure<br />
and the contribution to this activity of the metalbinding<br />
domains, carbohydrate domain and the C-terminal<br />
domains (residues 124-770) remain unknown. The APP<br />
N-terminal domain consists of nine β-strands and one<br />
α-helix which fold into a compact, globular domain. The<br />
helix represents the longest stretch of secondary structure,<br />
being 13 residues long.<br />
The overall aim of this project is to un<strong>der</strong>stand how<br />
monomer-to-dimer transition controls the activity and<br />
the metabolism of APP. We believe that the elucidation<br />
of biological regulatory mechanisms in APP can<br />
lead to the design of new approaches for manipulating<br />
the APP-system. This will help to better un<strong>der</strong>stand its<br />
function and to achieve new outcomes for a possible<br />
therapy and diagnosis.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(SFB 317 “<strong>Molekulare</strong> <strong>Biologie</strong> neuraler<br />
Mechanismen und Interaktionenen”, Graduiertenkolleg<br />
“<strong>Molekulare</strong> Zellbiologie”, Graduiertenkolleg<br />
“<strong>Molekulare</strong> und zelluläre Neurobiologie” and Projekt-’Sachbeihilfen’),<br />
from the BioRegion Rhein-<br />
Neckar (BMBF ), from the Alzheimer Forschung Initiative<br />
e.V. and the International Copper Association<br />
(ICA).<br />
PUBLICATIONS<br />
Moir, R.D., Lynch, T., Bush, A.I., Whyte, S., Henry,<br />
A., Portbury, S., Tanzi, R.E., Multhaup, G., Small,<br />
D.H., Beyreuther, K. and Masters, C.L. (1998). Relative<br />
increase in Alzheimer‘s disease of soluble forms<br />
of cerebral Aß amyloid protein precursor containing<br />
the kunitz protease inhibitory domain. J. Biol. Chem.<br />
273, 5013-9.<br />
Multhaup, G., Ruppert, T., Schlicksupp, A., Hesse, L.,<br />
Bill, E., Pipkorn, R., Masters, C.L. and Beyreuther, K.<br />
(1998). APP-copper complexes un<strong>der</strong>go site-specific<br />
fragmentation in the reduction of hydrogen peroxide.<br />
Biochemistry 37, 7224-7230.<br />
Ilg, T., Craik, D., Currie, G., Multhaup, G., and Bacic,<br />
A. (1998). Stage-specific proteophosphoglycan from<br />
leishmania mexicana amastigotes. Structural characterization<br />
of novel mono-, di-, and triphosphorylated<br />
phosphodiester-linked oligosaccharides. J. Biol.<br />
Chem. 273, 13509-23.<br />
Henry, A., Li, Q-X., Galatis, D., Hesse, L., Multhaup,<br />
G., Beyreuther, K., Masters, C.L. and Cappai, R.<br />
(1998). Inhibition of platelet activation by the Alzheimer‘s<br />
disease amyloid precursor protein. Br. J. Haematol.<br />
103, 402-415.<br />
Rossjohn, J., Cappai, R., Feil, S.C., Henry, A., McKinstry,<br />
W.J., Galatis, D., Hesse, L., Multhaup, G.,<br />
Beyreuther, K., Masters, C.L. and Parker, M.W.<br />
(1999). Crystal structure of the N-terminal, growth<br />
factor-like domain of Alzheimer‘s amyloid precursor<br />
protein. Nature Struc. Biol. 6, 327-331.<br />
Urban, S., Kruse, C. and Multhaup, G. (1999). A<br />
soluble form of the avian hepatitis B virus receptor.<br />
Biochemical characterization and functional analysis<br />
of the receptor ligand complex. J. Biol. Chem. 274,<br />
5707-5715.<br />
Beher, D., Elle, C., Un<strong>der</strong>wood, J., Davis, J.B., Ward,<br />
R., Karran, E., Roberts, G., Masters C.L., Beyreuther<br />
K. and Multhaup, G. (1999). Proteolytic fragments of<br />
Alzheimer’s disease-associated Presenilin 1 are present<br />
in synaptic organelles and growth cone membranes<br />
of rat brain. J. Neurochem. 72, 1564-1573.<br />
Multhaup, G., Hesse, L., Borchardt, T., Ruppert, T.,<br />
Cappai, R., Masters, C. L., and Beyreuther, K. (1999).<br />
Autoxidation of amyloid precursor protein and formation<br />
of reactive oxygen species. Adv. Exp. Med. Biol.<br />
448, 183-92.<br />
Huang, X., Atwood, C. S., Hartshorn, M. A., Multhaup,<br />
G., Goldstein, L. E., Scarpa, R. C., Cuajungco, M. P.,<br />
Gray, D. N., Lim, J., Moir, R. D., Tanzi, R. E., and<br />
Bush, A. I. (1999). The Abeta peptide of Alzheimer‘s<br />
disease directly produces hydrogen peroxide through<br />
metal ion reduction. Biochemistry 38, 7609-16.<br />
45
Multhaup, G., Hesse, L., Borchardt, T., Ruppert, T.,<br />
Cappai, R., Masters, C.L. and Beyreuther, K. (1999).<br />
Autoxidation of amyloid precursor protein and formation<br />
of reactive oxygen species. In Copper Transport<br />
and Its Disor<strong>der</strong>s: Molecular and Cellular Aspects,<br />
16, 183-192 (A. Leone and JFB Mercer, eds) Plenum,<br />
New York.<br />
Egensperger, R., Weggen, S., Ida, N., Multhaup, G.,<br />
Schnabel, R., Beyreuther, K., and Bayer, T. A. (1999).<br />
Reverse relationship between beta-amyloid precursor<br />
protein and beta-amyloid peptide plaques in Down‘s<br />
syndrome versus sporadic/familial Alzheimer‘s disease.<br />
Acta Neuropathol. (Berl) 97, 113-8.<br />
Multhaup, G., and Masters, C. L. (1999). Metal binding<br />
and radical generation of proteins in human neurological<br />
diseases and aging. Met. Ions Biol. Syst. 36,<br />
365-87.<br />
Lichtenthaler, S. F., Multhaup, G., Masters, C. L., and<br />
Beyreuther, K. (1999). A novel substrate for analyzing<br />
Alzheimer‘s disease gamma-secretase. FEBS Lett.<br />
453, 288-92.<br />
White, A.R., Reyes, R., Mercer, J.F.B., Camakaris, J.,<br />
Zheng, H., Bush, A.I., Multhaup, G., Beyreuther, K.,<br />
Masters, C.L. and Cappai, R. (1999). Copper levels<br />
are increased in the cerebral cortex and liver of APP<br />
and APLP2 knockout mice. Brain Res. 842, 439-444.<br />
Bayer, T. A., Cappai, R., Masters, C. L., Beyreuther,<br />
K., and Multhaup, G. (1999). It all sticks together - the<br />
APP-related family of proteins and Alzheimer‘s disease.<br />
Mol. Psychiatry 4, 524-528.<br />
Borchardt, T., Camakaris, J., Cappai, R., Masters, C.<br />
46<br />
L., Beyreuther, K., and Multhaup, G. (1999). Copper<br />
inhibits ß-amyloid production and stimulates the nonamyloidogenic<br />
pathway of amyloid precursor protein<br />
(APP) secretion. Biochem. J. 344, 461-467.<br />
White, A. R., Multhaup, G., Maher, F., Bellingham,<br />
S., Camakaris, J., Zheng, H., Bush, A. I., Beyreuther,<br />
K., Masters, C. L., and Cappai, R. (1999). The<br />
Alzheimer‘s disease amyloid precursor protein (APP)<br />
modulates copper-induced toxicity and oxidative<br />
stress in primary neuronal cultures. J. Neurosci. 19,<br />
9170-9179.<br />
Huang, X., Cuajungco, M.P., Atwood, C.S., Hartshorn,<br />
M.A., Tyndall, J.D.A., Hanson, G.R., Stokes,<br />
K.C., Leopold, M., Multhaup, G., Goldstein, L.E.,<br />
Scarpa, R.C., Saun<strong>der</strong>s, A.J., James Lim, Moir, R.D.,<br />
Glabe, C., Bowden, E.F., Masters, C.L., Fairlie, D.P.,<br />
Tanzi, R.E., and Bush, A.I. (1999). Cu(II) potentiation<br />
of Alzheimer Aß neurotoxicity. Correlation with cellfree<br />
hydrogen peroxide production and metal reduction.<br />
J. Biol. Chem. 274, 3711-6.<br />
Urban, S., Schwarz, C., Marx, U., Zentgraf, W. and<br />
Multhaup, G. (<strong>2000</strong>). Receptor recognition by a hepatitis<br />
B virus reveals a novel mode of high affinity<br />
virus-host interaction. EMBO J. 19, 1217-1227.<br />
Kulic, L., Walter, J., Multhaup, G., Teplow, D.B.,<br />
Romig, H., Capell, A., Steiner, H. and Haass, C.<br />
(<strong>2000</strong>). Separation of presenilin function in endoproteolysis<br />
of the ß-amyloid precursor protein and notch.<br />
Proc. Natl. Acad. Sci. USA, 97, 5913-5918.<br />
Borchardt, T., Schmidt, C., Camakaris, J., Cappai,<br />
R., Masters, C.L., Beyreuther, K. and Multhaup, G.<br />
(<strong>2000</strong>). Differential effects of zinc on amyloid precur-<br />
sor protein (APP) processing in copper-resistant variants<br />
of cultured Chinese hamster ovary cells. Cell.<br />
Mol. Biol, 46, 785-795.<br />
Capell, A., Steiner, H., Willem, M., Kaiser, H., Meyer,<br />
C., Walter, J., Lammich, S., Multhaup, G. and Haass,<br />
C. (<strong>2000</strong>). Maturation and pro-peptide cleavage of<br />
ß-secretase (BACE). J Biol Chem, (in press).<br />
THESES<br />
Dissertation<br />
Hesse, Lars (1998): Funktionelle Analyse von extrazellulären<br />
Domänen des Alzheimer Vorläuferproteins<br />
und an<strong>der</strong>er Mitglie<strong>der</strong> <strong>der</strong> APP-Genfamilie.<br />
Awards<br />
1998 ‚Award‘ <strong>der</strong> ‚International Copper Association‘<br />
(ICA)<br />
1999 Alzheimer Forschung Initiative (AFI) ‚Grant<br />
award‘<br />
STRUCTURE OF THE GROUP<br />
E-mail: g.multhaup@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Multhaup, Gerd, Priv.-<br />
Doz. Dr.<br />
Research associate Strausak. Daniel, Dr.*<br />
Postdoctoral fellows Hesse, Lars, Dr.*<br />
Schmidt, Carsten, Dr.*<br />
Ph.D. students Simons, Andreas*<br />
Strauss, Markus*<br />
Elle, Christine<br />
Techn. assistant Schlicksupp, Andrea<br />
*part of the time reported<br />
47
Herrmann Bujard<br />
I Expanding the Applicability of the<br />
Tet Regulatory Systems<br />
U. Baron, S. Freundlieb, R. Löw, Ch. Schirra-Müller<br />
Though very tight regulation of gene expression may<br />
be achieved with the Tet regulatory system - demonstrated<br />
by the successful control of the diphteria<br />
toxin gene in transgenic mice most impressively - it<br />
is sometimes difficult to initially establish cell lines or<br />
transgenic animals where genes encoding potentially<br />
toxic products are un<strong>der</strong> Tet control. The main reason<br />
for such failures is the transient state of the transferred<br />
DNA. During this period when chromatin suppression<br />
is missing, background expression from multiple<br />
copies of the target gene may be sufficient to kill<br />
the cell. We have deviced two approaches that help<br />
to overcome this limitaton. By fusing transcriptional<br />
silencing domains to the Tet repressor (TetR), tetracycline<br />
(Tc) controlled silencers (tTS) were developed<br />
that bind to the operator sequences within P tet (Fig. 1)<br />
48<br />
and shield the promoter from outside activation. Addition<br />
of doxycycline (Dox) will dissociate tTS from P tet<br />
and at the same time cause its activation via rtTA. The<br />
second approach makes use of gene transfer via retroviruses<br />
where single genomes can be delivered to<br />
the nucleus. Several vehicles including lentiviral vectors<br />
(collaboration with L. Naldini, Candiolo/Torino,<br />
Italy) were composed, which function well. Together,<br />
these components should not only abrogate present<br />
limitations, they offer themselves also for the development<br />
of tightly controllable gene delivery systems that<br />
may become useful in gene therapy. Several collaborations<br />
in this area are ongoing.<br />
Figure 1: The family of Tc-controlled regulatory proteins.<br />
TetR (red) was originally fused to a portion of the Herpes<br />
simplex virus activator protein VP16 (green) resulting in<br />
tTA (Tc controlled transactivator). Replacing the VP16<br />
moiety by multiples of 13 amino acid long minimal activation<br />
domains (upper left) has increased the specificity of<br />
tTA. A quadruple mutant of TetR exhibiting a reverse phenotype<br />
(violet) was fused to activation domains resulting in<br />
rtTA that requires Dox for operator binding. Modifying the<br />
DNA recognition specificity of tTA and rtTA (blue) as well<br />
as the dimerization surface of tTA (grey) led to tTA and rtTA<br />
versions that can be used to control two genes in a mutually<br />
exclusive way by Dox. Fusion of TetR with an altered<br />
dimerization surface to silencing domains led to Tc-controlled<br />
transcriptional silencers (tTS) that, in concert with<br />
rtTA, establish a repression/ activation system where in the<br />
„OFF“ state, i.e. in absence of Dox, P tet - a minimal promoter<br />
fused to an array of 7 tet operators - is shielded from<br />
outside activation.-<br />
Probing the sequence space of TetR<br />
T. Baldinger, U. Baron, M. Hasan, Ch. Schirra-Müller<br />
Most of the TetR mutants that have advantageously<br />
modified the Tet regulatory system resulted from powerful<br />
genetic approaches in E.coli. However, the phenotypes<br />
observed in the prokaryotic cell do frequently<br />
not correlate with those seen in the eukaryotic environment.<br />
Therefore, two eukaryotic genetic systems<br />
were conceived; one developed by W. Hillen and his<br />
group (Universität Erlangen) is based on S.cerevisiae,<br />
the other on mammalian cells. With these systems,<br />
the sequence space of fusions between TetR and functional<br />
domains can be directly explored e.g. by screening<br />
or selecting for activation or silencing of marker<br />
functions. First screens revealed a number of interesting<br />
TetR mutants of which one - identified in the lab in<br />
Erlangen - exhibits a striking reverse phenotype that<br />
is superior to the originally described rtTA. Furthermore,<br />
we are interested in whether tTA/rtTA mutants<br />
can be identified which discriminate between different<br />
tetracycline <strong>der</strong>ivatives or which may even be susceptible<br />
to non-tetracycline compounds as inducers.<br />
If successful, this search could lead to well defined<br />
and largely homologous regulatory circuits that would<br />
allow to control several genes independently from<br />
each other by just using different inducer molecules<br />
(collaboration with W. Hillen and B. Berkhout).<br />
Controlling genes in vivo<br />
S. Berger, M. Hasan, R. Löw, K. Schönig<br />
Our transgenic mouse lines that synthesize tTA or<br />
rtTA specifically in hepatocytes and in specific areas<br />
of the brain, respectively, were used to measure kinet-<br />
ics of induction and shut-off of gene activities in these<br />
two compartments of the animal. Using a non-invasive<br />
approach that monitores luciferase activity in live<br />
animals, repeated cycles of gene activation and deactivation<br />
can be followed in individual mice (Fig. 2).<br />
These studies have provided not only insights into the<br />
time course of switching between active and inactive<br />
states of a gene via Dox in live animals but also into<br />
the longevity of the expression system. Thus, independent<br />
of whether the luciferase reporter gene was kept<br />
in the „ON“ or in the „OFF“ state, regulatory cycles<br />
could be re-initiated after 6 months in all individuals<br />
tested. As any target gene may be coregulated together<br />
with the luciferase gene as indicator via the bidirectional<br />
P tet promoters, monitoring luciferase activity in<br />
live animal will be indicative for the activity of the<br />
gene un<strong>der</strong> study (Fig. 2). In summary, the newly<br />
developed rtTA‘s together with some novel doxycycline<br />
<strong>der</strong>ivatives that are presently being characterized<br />
and the induction studies have made us confident that<br />
genes of interest can be controlled in the mouse brain<br />
tightly and reliably over long periods of time. In<br />
this context, it is our aim to control gene functions<br />
indirectly by interferring at the level of transcription<br />
initiation and/or in post-transcriptional processes, an<br />
approach which would leave the endogeneous gene<br />
of interest untouched in its genomic setting. Therefore,<br />
we explore several experimental options which<br />
include padlock RNA‘s, ribozymes and zinc finger<br />
proteins. We have chosen the NRI subunit of the<br />
NMDA receptor as primary target for this work. Our<br />
technology is used in several rather challenging collaborations<br />
to generate animal models for human diseases.<br />
One exciting example is a mouse model for<br />
prion disease that is being developed by S. Prusiner<br />
and P. Tremblay, UCSF, San Francisco, USA.<br />
49
Figure 2: Monitoring<br />
gene activation and<br />
inactivation in live animals.<br />
Mice double transgenic<br />
for tTA and the<br />
luciferase gene un<strong>der</strong><br />
Ptet control are injected<br />
with luciferin and anesthesized<br />
for 5 min.<br />
before they are placed<br />
into the Hamamatsu<br />
photon counting device<br />
Argus 20 (left upper part<br />
of fig.). Photons emitted<br />
by luciferase active in<br />
liver (left most animal)<br />
and in brain (right<br />
animal) are collected as<br />
imaged below. The single<br />
transgenic animal in the<br />
middle serves as control.<br />
Feeding Dox to the<br />
“liver mouse” abolishes<br />
luciferase activity within<br />
5 days. Removal of Dox<br />
in the drinking water reestablishes<br />
luciferase activity within 5 days (“brain mouse” as control). The cycle of switching was repeated with the same<br />
animals after 3 months with identical results, except that 2 days of exposure to Dox were sufficient to shut off luciferase activity<br />
in the liver. The luciferase gene is driven by a bidirectional Ptet which coregulates the Cre recombinase gene. Luciferase<br />
activity correlates with Cre activity. Whole body images of the animals are shown.<br />
II The Merozoite Surface Protein 1 of<br />
the Human Malaria Parasite Plasmodium<br />
falciparum<br />
Malaria is one of the most widely spread infectious<br />
diseases with around 40 % of the world‘s population<br />
living in areas at risk. The hope for an effective vaccine<br />
candidate against infection of P.falciparum, the<br />
most lethal one among the human malaria parasites,<br />
relies on the subunit concept where individual components<br />
of the parasite are utilized to elicit a protective<br />
immune response. Our studies focus on a 190 kDa<br />
protein which is the major surface protein of the mero-<br />
50<br />
zoite (MSP-1), one of the parasite‘s blood forms. This<br />
protein is believed to play a role during invasion of<br />
erythrocytes by the parasite. It is a target of the human<br />
immune response at the humoral as well as at the cellular<br />
level. Such findings suggest that MSP-1 may<br />
elicit a protective immune response when used as a<br />
vaccine in humans. The aim of our work is to develop<br />
an experimental vaccine that is suitable for human<br />
trials. Moreover, we are interested in the function of<br />
MSP-1 and its structure at the parasite‘s surface.<br />
Synthetic msp-1 genes provide material for<br />
immunological, structural and functional studies<br />
C. Epp, C. Fernandez-Becerra, C. Schmid, I. Türbachova,<br />
N. Westerfeld<br />
P.falciparum DNA has an exceptionally high AT content<br />
which amounts to 76 % in the coding area of<br />
MSP-1. The high AT content has prevented the stable<br />
cloning of large genes of this parasite and thus severely<br />
hampered their study. We have synthesized the genes<br />
of both MSP-1 prototypes (together around 10 000<br />
bp) based on human codon frequencies and are now in<br />
a position to produce MSP-1 and <strong>der</strong>ivatives thereof<br />
in various heterologous systems. During maturation<br />
of merozoites, MSP-1 un<strong>der</strong>goes proteolytic cleavage<br />
resulting in five major fragments that can be isolated<br />
as one complex from the merozoite surface. The proteolytic<br />
cleavage sites were reconciled in the design of<br />
our synthetic genes, and accordingly expression vehicles<br />
encoding the various fragments are available as<br />
well. Efficient expression of the intact genes as well<br />
as of DNA encoding the processing products has been<br />
achieved in E.coli. Full size MSP-1 is also produced<br />
and secreted into the medium by bacterial L-forms<br />
(collaboration with M. Kujau, IMB, Jena) from where<br />
it can be isolated in soluble form. We now concentrate<br />
on devicing a production procedure for MSP-1<br />
that is suitable for „good manufacturing practice“.<br />
Our highly purified preparations will also be used<br />
for structural studies and for exploring interactions<br />
between the individual processed domains as well as<br />
between MSP-1 and the erythrocyte surface. A second<br />
focus of this project is the integration of msp-1 into<br />
viral systems that are suitable as vaccine carriers in<br />
humans, such as vaccinia and measles viruses.<br />
A MSP-1 based ELISA for sero-epidemiological<br />
surveys<br />
C. Epp, I. Idler, I. Türbachova, S. Weerasuriya<br />
The availability of the various MSP-1 processing products<br />
in apparently native conformation has enabled us<br />
to develop an ELISA that can be used for serum analysis.<br />
In a first approach, we have examined the sera<br />
from two trials in which Aotus monkeys were immunized<br />
with MSP-1 and challenged with merozoites<br />
(collaboration with S. Herrera, Cali, Colombia). Full<br />
protection was achieved in 60 % of the animals. Interestingly,<br />
protection correlated well with the humoral<br />
response against certain areas of MSP-1. Using the<br />
new ELISA, we are now re-examining the sera collected<br />
during our earlier epidemiological studies in<br />
West Africa. Our previous analysis has indicated a<br />
correlation between antibody titers towards certain<br />
regions of MSP-1 and a reduced risk of reinfection<br />
by P.falciparum.We hope that our new analysis will<br />
reveal more clearly which regions of MSP-1 may be<br />
involved in eliciting a protective humoral response.<br />
The ELISA may eventually permit the development of<br />
a diagnostic tool with predictive properties.<br />
Attempts to identify interactions between<br />
MSP-1 and the surface of erythrocytes<br />
P. Burghaus, I. Türbachova<br />
As the major protein at the surface of merozoites,<br />
MSP-1 has been implicated in early association events<br />
during the erythrocyte invasion. Following the successful<br />
strategy described for the Duffy binding protein,<br />
we have exposed full length MSP-1 as well as<br />
stepwise truncated versions of the protein on the surface<br />
of HeLa cells, fixed, as in the parasite, by a<br />
51
GPI-anchor. Moreover, in collaboration with D. Soldati<br />
(<strong>ZMBH</strong>), we have generated several recombinant<br />
Toxoplasma gondii strains that also expose at the<br />
surface full size MSP-1, the p42 or the p19 processing<br />
product, respectively. In both, the HeLa cell and<br />
the T.gondii system, monoclonal antibodies directed<br />
towards conformational epitopes of MSP-1 interact<br />
efficiently with the protein at the surface of the respective<br />
cells. Nevertheless, so far we were neither able<br />
to unequivocally demonstrate an interaction between<br />
MSP-1 and erythrocytes nor can we definitely rule out<br />
such interactions. The failure to reveal the putative<br />
affinity may be due to a number of reasons. We, therefore,<br />
will further pursue these studies.<br />
External Funding<br />
During the period reported, our research was supported<br />
by grants from the BMBF (Forschungsschwerpunkt<br />
Tropenmedizin), the Volkswagen-Stiftung, the<br />
Deutsche Forschungsgemeinschaft (SFB 544 “Kontrolle<br />
tropischer Infektionskrankheiten” and Projekt-<br />
’Sachbeihilfen’), from the EU, the BioRegion Rhein-<br />
Neckar (BMBF - Fa. Knoll AG) and the Fonds <strong>der</strong><br />
Chemischen Industrie Deutschlands.<br />
PUBLICATIONS<br />
Tremblay, P., Meiner, Z., Galou, M., Heinrich, C.,<br />
Petromilli, C., Lisse, T., Cayateno, J., Torchia, M.,<br />
Mobley, W., Bujard, H., DeArmond, S.J. and Prusiner,<br />
S.B. (1998). Doxycycline control of prion protein<br />
transgene expression modulates prion disease in mice.<br />
Proc. Natl. Acad. Sci. USA 95, 12580-12585.<br />
Baron, U., Schnappinger, D., Helbl, V., Gossen, M.,<br />
Hillen, W. and Bujard, H. (1999). Generation of conditional<br />
mutants in higher eukaryotes by switching<br />
52<br />
between the expression of two genes. Proc. Natl.<br />
Acad. Sci. USA 96, 1013-1018<br />
Freundlieb, S., Schirra-Müller, C. and Bujard, H.<br />
(1999). A tetracycline controlled activation /repression<br />
system for mammalian cells. J. Gene Med. 1, 4-12.<br />
Pan, W., Ravot, E., Tolle, R., Frank, R., Mosbach, R.,<br />
Türbachova, I. and Bujard, H. (1999). Vaccine candidate<br />
MSP-1 from Plasmodium falciparum: a redesigned<br />
4917 bp polynucleotide enables synthesis and<br />
isolation of full length protein from E.coli and mammalian<br />
cells. Nucl. Acids Res. 27, 1094-1103.<br />
Redfern, C.H., Coward, P., Degtyarev, M.Y., Lee, E.K.,<br />
Kwa, A., Hennighausen, L., Bujard, H., Fishman, G.I.<br />
and Conklin, B.R. (1999). In vivo conditional expression<br />
and signaling of a specifically designed G i -coupled<br />
receptor. Nature Biotech. 17, 165-169.<br />
Burghaus, P.A., Gerold, P., Pan, W., Schwarz, R.T.,<br />
Lingelbach, K. and Bujard, H. (1999). Analysis of<br />
recombinant merozoite surface protein-1 of Plasmodium<br />
falciparum expressed in mammalian cells. Mol.<br />
Biochem. Parasitol. 104, 171-183.<br />
Lavon, I., Goldberg, I., Amit, S., Landsman, L., Jung,<br />
S., Tsuberi, B., Barshack, I., Kopolovic, J., Galun, E.,<br />
Bujard, H. and Ben-Neriah, Y. (<strong>2000</strong>) High susceptibility<br />
to bacterial infection, but no liver dysfunction,<br />
in mice compromised for hepatocyte NF-κB activation.<br />
Nat. Med. 6, 573-577.<br />
Baron, U. and Bujard, H. (<strong>2000</strong>) The Tet repressor<br />
based system for regulated gene expression in eukaryotic<br />
cells: principles and advances. Methods Enzymol.<br />
327, 659-686.<br />
Mansuy, I. and Bujard, H. (<strong>2000</strong>) Tetracycline-regulated<br />
gene expression in the brain. Curr. Op. Neurobiol.,<br />
in press.<br />
Urlinger, S., Baron, U., Thellmann, M., Hasan, M.,<br />
Bujard, H. and Hillen, W. (<strong>2000</strong>). Exploring the<br />
sequence space for tetracycline dependent transcriptional<br />
activators: novel mutations yield expanded<br />
range and sensitivity. Proc. Natl. Acad. Sci. USA, 97,<br />
7963-68.<br />
THESES<br />
Diploma<br />
Epp, Christian (1998): Analyse <strong>der</strong> humoralen Immunantwort<br />
von Aotus-Affen gegen eine Immunisie–<br />
rung mit MSP-1 aus Plasmodium falciparum<br />
Schönig, Kai (1998): Konstruktion und Analyse von<br />
„integrierten Regulationseinheiten“ zur Tetrazyklin<br />
kontrollierten Genexpression in Säugerzellen<br />
Schmid, Christina (1999): Untersuchungen zum Oberflächenprotein<br />
1 von Merozoiten (MSP-1) des Mala–<br />
riaerregers Plasmodium falciparum: Kontrollierte Synthese<br />
des 42kDa-Fragments in Säugerzellen<br />
Dissertation<br />
Baron, Udo (1998): Weiterentwicklung <strong>der</strong> Methodik<br />
<strong>der</strong> Tetrazyklin-kontrollierten Genexpression zur Ana–<br />
lyse <strong>der</strong> Funktion von Genen in komplexen eukaryotischen<br />
Systemen.<br />
Awards<br />
Ruperto Carola Preis 1998 <strong>der</strong> Universität Heidelberg,<br />
for outstanding PhD Thesis to Udo Baron<br />
STRUCTURE OF THE GROUP<br />
E-mail: bujardh@sun0.urz.uni-heidelberg.de<br />
Group lea<strong>der</strong> Bujard, Hermann, Prof. Dr.<br />
Guest scientist del Portillo, Hernando, Prof. Dr.*<br />
Research<br />
Associate Freundlieb, Sabine, Dr.<br />
Postdoctoral Baron, Udo, PhD<br />
fellows Fernandez-Becerra, Carmen, PhD*<br />
Hasan, Mazahir, PhD*<br />
Löw, Rainer, PhD*<br />
Weerasuriya, Siromi, PhD*<br />
Ph.D. students Berger, Stefan, Dipl.Chem.<br />
Epp, Christian, Dipl.Biol.<br />
Idler, Irina, Dipl.Biol.*<br />
Kolster, Kathrin, Dipl.Biol.*<br />
Schönig, Kai, Dipl.Biol.<br />
Türbachova, Ivana, Dipl.Biol.<br />
Westerfeld, Nicole, Dipl.Biol.<br />
Diploma<br />
students Baldinger, Tina*<br />
Schmid, Christina*<br />
Techn. assistants Rittirsch, Melanie*<br />
Schirra-Müller, Christiane<br />
Wahedy, Soraya*<br />
* part of the time reported<br />
53
Christine Clayton<br />
Molecular Cell Biology of Trypanosomes<br />
Introduction<br />
Salivarian trypanosomes are unicellular parasites that<br />
live in the blood and tissue fluids of mammals. They<br />
cause sleeping sickness in humans and also infect<br />
domestic animals and wildlife, mainly in subSaharan<br />
Africa. The geographical restriction is determined by<br />
the fact that the parasites are usually transmitted from<br />
one mammal to the next by Tsetse flies. They develop<br />
and multiply within the insect midgut and can un<strong>der</strong>go<br />
a sexual cycle before infection of another mammal<br />
via the insect salivary glands. Salivarian trypanosomes<br />
do not multiply inside cells, unlike the closely<br />
related tropical disease parasites Trypanosoma cruzi<br />
and Leishmania. At present about 300 000 people are<br />
infected with the sleeping sickness trypanosomes Trypanosoma<br />
brucei rhodesiense and Trypanosoma gambiense.<br />
The disease is invariably fatal unless treated,<br />
and very few drugs are available, especially to combat<br />
the late stages of the disease when the parasites invade<br />
the brain. Moreover, most of the available drugs are<br />
either too expensive or unacceptably toxic and drug<br />
resistance is developing.<br />
Trypanosomes and Leishmanias belong to the genus<br />
Kinetoplastida, which branched extremely early in<br />
eucaryotic evolution. As a consequence they exhibit<br />
a number of remarkable deviations from standard<br />
eucaryotic paradigms. In the realm of gene expression,<br />
most transcription is polycistronic, all mRNAs are<br />
trans spliced, there is almost no developmental regulation<br />
of transcription, and the mitochondrial RNAs<br />
are extensively edited. Energy and redox metabolism<br />
also exhibit unique features: for example, the glycolytic<br />
enzymes are compartmentalised in an organelle,<br />
54<br />
Figure 1: Electron micrographs of trypanosomes. On the left<br />
are normal trypanosomes with many round glycosomes (Gly).<br />
On the right are cells with a 90% reduction in PEXII (Evelyn<br />
Baumgart, Anatomy Dept.).<br />
the glycosome. At the same time, the parasites show<br />
many conserved features. For example, the glycosome<br />
is a specialised type of peroxisome, and trans splicing<br />
is mechanistically related to cis splicing.<br />
Our work concentrates on Trypanosoma brucei brucei,<br />
a close relative of the sleeping sickness trypanosomes<br />
which is not infective for humans. The parasites grow<br />
in laboratory rodents or in vitro and life-cycle related<br />
differentiation can be mimicked in culture. They are<br />
diploid, carrying about 60 Mb of DNA on 11 chromosome<br />
pairs; a sequencing project is in progress. A full<br />
palette of genetic manipulation methods is available,<br />
including gene replacement by homologous recombination<br />
and inducible gene expression. Conditional<br />
knockouts can be made by placing the gene of interest<br />
un<strong>der</strong> control of a tetracycline-inducible promoter,<br />
then deleting the remaining gene copies via homologous<br />
recombination.<br />
In our research on trypanosomes, we try to keep two<br />
aims in view. Firstly, we concentrate on aspects of<br />
the parasites that uniquely distinguish them from their<br />
mammalian hosts, and may therefore represent suitable<br />
targets for specific anti-parasitic chemotherapy.<br />
Secondly, by examining the similarities and differences<br />
between fundamental processes in trypanosomes,<br />
mammals and yeast we can clearly distinguish<br />
those aspects that are conserved throughout eucaryotic<br />
evolution and therefore likely to be essential from<br />
those that are required for specialised lifestyles.<br />
Control of gene expression<br />
Maciej Drodzd, Antonio Gonzales, Claudia Hartmann,<br />
Henriette Irmer, Luis Quilada<br />
Specialised surface molecules are essential for the<br />
survival and virulence of pathogens. In the case of<br />
trypanosomes, the forms that grow in the mammal<br />
(bloodstream forms) wear a uniform coat of variant<br />
surface glycoprotein (VSG), expressed from a single<br />
gene chosen from a repertoire of up to 1000 different<br />
VSG genes. The VSG expressed can be changed either<br />
through recombination or by transcriptional switches.<br />
This enables the parasites to change their surface coat,<br />
thereby evading the immune response. In the Tsetse<br />
midgut, the VSG is replaced by the EP and GPEET<br />
repetitive, acidic proteins. All these surface proteins<br />
are retained on the plasma membrane by a glycosyl<br />
phosphatidylinositol (GPI) lipid anchor.<br />
Normally, protein-coding genes can only be functionally<br />
expressed by RNA polymerase II. This is because<br />
the mRNAs have to be modified at the 5‘-end by<br />
enzymes that are associated exclusively with RNA<br />
polymerase II. The Kinetoplastids are not subject to<br />
this restriction because the 5‘ spliced lea<strong>der</strong> is capped.<br />
The VSG and the EP/GPEET genes are transcribed<br />
by RNA polymerase I as part of polycistronic tran-<br />
scription units. Nearly all other protein-coding genes<br />
are transcribed by RNA polymerase II, but again transcription<br />
is polycistronic. Although the expression of<br />
many genes has to be strongly regulated to enable<br />
survival in the disparate environments of Tsetse and<br />
mammal, there is no evidence for any developmental<br />
control of RNA polymerase II transcription. Instead,<br />
regulation usually requires sequences which are found<br />
in the 3‘-untranslated regions and control RNA degradation<br />
and/or translation. Polymerase I-mediated transcription<br />
of the VSG genes is, exceptionally, subject to<br />
strong developmental regulation, but the EP/GPEET<br />
gene transcription is only weakly regulated.<br />
To compensate for the lack of transcriptional regulation,<br />
the EP/GPEET mRNAs are extremely unstable<br />
and very poorly translated in bloodstream forms,<br />
so that no protein product is detectable. A uridinerich<br />
26 nt sequence in the 3‘-untranslated region of<br />
EP/GPEET mRNA is essential for this control. We<br />
have examined the mechanism of RNA degradation<br />
in detail. Relatively stable RNAs such as the actin<br />
mRNA appear to be degraded by the standard mechanism<br />
seen in eucaryotes: initial destruction of the<br />
poly(A) tail at the 3‘-end is followed by degradation<br />
of the rest of the mRNA. The EP/GPEET mRNAs<br />
are in contrast rapidly degraded in bloodstream forms<br />
without prior degradation of the poly(A) tail. Detailed<br />
characterisation of the EP RNA secondary structure<br />
indicated that the 26mer is in an exposed, extended<br />
conformation, so it should be accessible to RNA-binding<br />
proteins or specific endonucleases. Searches for<br />
specific 26mer-binding proteins by several methods<br />
have however proved fruitless.<br />
Searches of the available genome sequence have<br />
revealed a number of potential trypanosome proteins<br />
that are very similar to yeast enzymes involved in<br />
mRNA degradation and processing of stable RNAs, or<br />
55
to proteins involved in regulation of RNA processing.<br />
The relevant trypanosome genes and proteins are currently<br />
being functionally characterised in or<strong>der</strong> to pin<br />
down their roles in regulated and constitutive RNA<br />
degradation.<br />
The Glycosome<br />
Christina Guerra-Giraldez, Sandra Helfert, Alexan<strong>der</strong><br />
Maier, Sebastian Schulreich<br />
Bloodstream trypanosomes compartmentalise the first<br />
nine enzymes of glucose and glycerol metabolism in<br />
a peroxisome-related microbody, the glycosome. Glucose<br />
is metabolised mostly to pyruvate, with a very<br />
low energy yield of only 2 molecules of ATP per molecule<br />
of glucose. In procyclic trypanosomes, the major<br />
energy sources are amino acids; although glycolysis<br />
is still present, the products are much more efficiently<br />
used in a well-developed mitochondrion, yielding up<br />
to ten times as much ATP.<br />
Normally, bloodstream trypanosome glycolysis involves<br />
an oxidation step. To maintain this oxidation,<br />
the parasites have two alternatives. Un<strong>der</strong> aerobic conditions<br />
they can indirectly use oxygen, via a mitochondrial<br />
oxidase. Alternatively, at least in short-term<br />
incubations, they can recover oxidising power by a<br />
corresponding reduction of triose phosphate, converting<br />
each molecule of glucose to one molecule each of<br />
pyruvate and glycerol, generating only one molecule<br />
of ATP per glucose. It has therefore generally been<br />
assumed that bloodstream trypanosomes can survive<br />
anaerobically. By combining reverse genetics with the<br />
use of inhibitors, we have found that trypanosomes<br />
cannot survive anaerobically. This opens up new possibilities<br />
for chemotherapy.<br />
The compartmentation of glycolysis in a microbody<br />
is unique in evolution. We would like to know which<br />
(if any) advantages this compartmentation affords.<br />
56<br />
We have therefore been attempting to create conditional<br />
mutants in glycosome membrane biogenesis.<br />
The overall strategy is to identify genes encoding<br />
glycosomal membrane proteins, then to use tetracycline-<br />
inducible gene expression to make a conditional<br />
knockout. The first candidates for this approach were<br />
the two most abundant proteins of the glycosomal<br />
membrane. The first to be studied, a 24kD protein, is<br />
the trypanosome homologue of yeast Pex11p, which<br />
is required for peroxisome division. The trypanosome<br />
PEX11 gene complements the yeast pex11∆<br />
deletion mutant, and parasites with reduced levels of<br />
PEX11 have fewer, larger glycosomes than normal.<br />
The second protein, GIM5, has a mass of 26kD and<br />
shows no significant similarity to any other proteins<br />
in the databases (apart from two predicted proteins<br />
from the closely-related parasite Leishmania). Experiments<br />
with conditional knockouts have demonstrated<br />
that GIM5 is essential for parasite survival, but have<br />
not yet given any indication of its precise function.<br />
The protein spans the membrane twice, with both termini<br />
exposed on the cytosolic side, and forms dimers<br />
in vivo.<br />
Because glycosomes are related to peroxisomes and<br />
the mechanism of organelle biogenesis is conserved,<br />
we can also find genes involved in glycosome biogenesis<br />
by homology. One candidate for this approach is<br />
the trypanosome PEX2 gene. PEX2 is essential for<br />
peroxisome membrane biogenesis in yeast. The generation<br />
of conditional knockouts is in progress.<br />
The secretory pathway<br />
Iris Ansorge, Michael Herberger, Alexan<strong>der</strong> Maier<br />
The secretory pathway of kinetoplastids is similar to<br />
that of other eucaryotes. The predominance of GPIanchored<br />
surface molecules is however a specialisa-<br />
tion that appears to be unique to primitive unicellular<br />
organisms. Although a few transmembrane proteins<br />
have been identified, the vast majority of trypanosome<br />
surface proteins are GPI-anchored. Even the transferrin<br />
receptor is a heterodimer that is completely unrelated<br />
to mammalian transferrin receptors; one of the<br />
subunits, ESAG6, is GPI-anchored, while its partner,<br />
ESAG7, lacks any direct membrane attachment. The<br />
anchors of bloodstream trypanosomes contain 14-carbon<br />
lipids so are shorter than the lipids on mammalian<br />
anchors. In mammalian cells, GPI-anchored proteins<br />
partition into lipid microdomains („rafts“) that<br />
are poorly soluble in Triton X-100 and may be important<br />
in selective trafficking of GPI-anchored proteins<br />
to particular parts of the plasma membrane. The trypanosome<br />
plasma membrane has three specialised<br />
domains: the flagellar membrane, the rest of the surface,<br />
and an inwardly-directed pocket at the base of<br />
the flagellum which is the only site of membrane trafficking<br />
(the flagellar pocket). Some proteins (such<br />
as VSG) cover the entire surface while others are<br />
restricted to one or two domains. Collaborative experiments<br />
demonstrated that VSG, which forms homodimers,<br />
partitions poorly (50%) into the detergent-insoluble<br />
fraction, whether it is integrated into the trypanosome<br />
membrane or in artificial lipid bilayers. The<br />
heterodimeric transferrin receptor, which is normally<br />
restricted to the membrane and matrix of the flagellar<br />
pocket, is not retained at all in detergent-insoluble<br />
fractions, perhaps because only a single GPI anchor<br />
per dimer is available.<br />
Coatomer is a complex of seven proteins (α, β, γ, δ,<br />
ε and ζ –COP) that is involved in the formation of vesicles<br />
that move between the endoplasmatic reticulum<br />
and the Golgi apparatus. Work in Heidelberg and elsewhere<br />
has yielded results suggesting that the endoplasmatic<br />
reticulum and coatomer are involved in peroxisome<br />
biogenesis. The C-terminus of mammalian<br />
PEX11 includes a coatomer-binding sequence. To find<br />
out whther coatomer-binding by PEX11 was conserved<br />
throughout evolution, we tested the interaction<br />
of the relevant PEX11 sequences with coatomer from<br />
trypanosomes, yeast and mammals. A necessary preliminary<br />
was the partial characterisation of trypanosome<br />
coatomer. We found that that the specificity of<br />
coatomer binding is conserved throughout evolution.<br />
Trypanosome PEX11 binds coatomer, but mutants<br />
that lack binding are still functional in vivo. Yeast<br />
PEX11 does not have a C-terminal coatomer-binding<br />
sequence. Thus binding of coatomer by the C-terminus<br />
of PEX11 is not functionally essential.<br />
Redox metabolism and drug resistance<br />
Stephan Krieger, Sanjay Shahi, Claudia Hartmann<br />
Like all other cells, trypanosomes require a reducing<br />
environment in their cytosol, and contain a number of<br />
cofactors and proteins that act to maintain the correct<br />
redox balance. In nearly all cells from bacteria to man,<br />
one of these cofactors is glutathione. The thiol group<br />
of glutathione is maintained in the reduced state by<br />
glutathione reductase. In trypanosomes, most of the<br />
glutathione is found in the form of trypanothione (two<br />
glutathione moieties conjugated to spermidine). Trypanothione<br />
is maintained in the dithiol state by a<br />
highly specific enzyme, trypanothione reductase. The<br />
uniqueness of this system makes it a strong candidate<br />
for targeted drug design. To test the precise role of<br />
trypanothione reductase in vivo, we created trypanosomes<br />
with an inducible knockout. Trypanothione<br />
reductase was found to be essential for trypanosome<br />
growth both in vitro and in a mammalian host. This<br />
was the first time that an inducible knockout parasite<br />
had been tested in an animal model system. We found<br />
that at least 90% inhibition of trypanothione reductase<br />
will be required for effective chemotherapy. Parasites<br />
57
with low levels of the enzyme were highly susceptible<br />
to oxidative stress.<br />
A variety of drugs is excreted from cancer cells in the<br />
form of glutathione-drug conjugates by ABC transporters<br />
of the MRP (multidrug resistance protein)<br />
family. In some Leishmania species, arsenite and antimonial<br />
drugs can be sequestered as conjugates with<br />
trypanothione. The only drugs available against latestage<br />
trypanosomiasis in east Africa are arsenical<br />
<strong>der</strong>ivatives, but resistance to these drugs is emerging.<br />
To find out if MRP-like trypanothione-arsenical conjugate<br />
pumps might play a role in resistance, we have<br />
cloned and sequenced trypanosome homologues of<br />
two Leishmania MRP-like genes. Cells over-expressing<br />
the transporters will be characterised with respect<br />
to drug sensitivity and other characteristics.<br />
Metabolic compartmentation in Toxoplasma<br />
Martina Ding, Frank Voncken<br />
D. Soldati (<strong>ZMBH</strong>) and I became intrigued by the<br />
complete absence of reports of peroxisomes in malaria<br />
parasites and related organisms such as Toxoplasma. A<br />
partial sequence for catalase (a peroxisomal marker)<br />
was identified in the Toxoplasma sequence database.<br />
The predicted protein has all concensus amino acids<br />
for enzyme activity, and a C-terminal peroxisomal<br />
targeting signal. To our surprise, immunofluorescent<br />
labelling of Toxoplasma with antibodies to two different<br />
anti-peptide antibodies gave predominantly cytosolic<br />
labelling, and the addition of the targeting signal<br />
to the green flourescent protein also resulted in cytosolic<br />
flourescence. Cell fractionations gave similar<br />
results. Thus so far we have no evidence that Toxoplasma<br />
contains peroxisomes.<br />
58<br />
Collaborations<br />
Dr. Luise Krauth-Siegel (Biochemie-<strong>Zentrum</strong>, Heidelberg)<br />
– trypanothione reductase.<br />
Dr. Dietmar Steverding (Parasitologie, Heidelberg) –<br />
transferrin receptor.<br />
Dr. Martina Bremser and Prof. F. Wieland (Biochemie-<strong>Zentrum</strong>,<br />
Heidelberg) - coatomer.<br />
Dr. Dominique Soldati (<strong>ZMBH</strong>) – Toxoplasma peroxisomes<br />
Drs. Jürgen Benting and Kai Simons, EMBL (rafts)<br />
Dr. Paul Michels (Institute for Cellular Pathology,<br />
Brussels, Belgium) - glycosomes and glycolysis.<br />
Prof. Kenneth Stuart (Seattle Biomedical Research<br />
Institute, Washington, Seattle) and Prof. Ullrich<br />
Goeringer (Universität Darmstadt) – inducible knockouts<br />
of genes involved in RNA editing.<br />
Dr. Roland Kaminsky and Dr. Pascal Maser (Tropeninstitut,<br />
Basel) - drug resistance.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(SFB 352 “<strong>Molekulare</strong> Mechanismen intrazellulärer<br />
Transportprozesse”, SFB 544 “Kontrolle tropischer<br />
Infektionskrankheiten”, Graduiertenkolleg “Kontrolle<br />
<strong>der</strong> Genexpression in pathogenen Mikroorganismen”<br />
and Projekt-’Sachbeihilfen’) from the Human Frontier<br />
Science Programme Organization, the EMBO, the<br />
DAAD, the BMBF (Forschungsschwerpunkt Tropenmedizin)<br />
and from the Fonds <strong>der</strong> Chemischen Industrie.<br />
PUBLICATIONS<br />
Original articles<br />
Ansorge, I., Steverding, D., Melville, S., Hartmann,<br />
C., and Clayton, C. (1999). Transcription of ‘inactive’<br />
expression sites in African trypanosomes leads<br />
to expression of multiple transferrin receptor RNAs<br />
in bloodstream forms. Mol. Biochem. Parasitol. 101,<br />
81-94.<br />
Benting, J., Rietveld, A., Ansorge, I., and Simons, K.<br />
(1999). Acyl and alkyl chain length of GPI anchors is<br />
critical for raft association in vitro. FEBS Lett. 462,<br />
47-50.<br />
Blattner, J., Helfert, S., Michels, P., and Clayton,<br />
C. E. (1998). Compartmentation of phosphoglycerate<br />
kinase in Trypanosoma brucei plays a critical role<br />
in parasite energy metabolism. Proc. Natl. Acad. Sci.<br />
USA 95, 11596-11600.<br />
Clayton, C.E., Ha, S., Rusche, L., Hartmann, C.,<br />
Beverley, S. M. (<strong>2000</strong>). Tests of heterologous promoters<br />
and intergenic regions in Leishmania major. Mol.<br />
Biochem. Parasit. 105, 163-167.<br />
Ding, M., Clayton, C.E. and Soldati, D. (<strong>2000</strong>). Toxoplasma<br />
gondii catalase: are there peroxisomes in Toxoplasma?<br />
. J. Cell Sci. 105, (in press).<br />
Drozdz, M., and Clayton, C. E. (1999). Confirmation<br />
of a regulatory 3‘-untranslated region from Trypanosoma<br />
brucei. RNA 5, 1632-1644.<br />
Hartmann, C., Hotz, H.-R., McAndrew, M., and Clayton,<br />
C. (1998). Effect of multiple downstream splice<br />
sites on polyadenylation in Trypanosoma brucei. Mol.<br />
Biochem. Parasitol. 93, 149-152.<br />
Hotz, H.-R., Biebinger, S., Flaspohler, J., and Clayton,<br />
C. E. (1998). PARP gene expression: regulation at<br />
many levels. Mol. Biochem. Parasitol. 91, 131-143.<br />
Krieger, S., Schwarz, W., Ariyanagam, M.R., Fairlamb,<br />
A., Krauth-Siegel, L., and Clayton, C. E. (<strong>2000</strong>).<br />
Trypanosomes lacking trypanothione reductase are<br />
avirulent and show increased sensitivity to oxidative<br />
stress. Mol. Microbiol. 35, 542-552.<br />
Lorenz, P., Meier, A., Erdmann, R., Baumgart, E., and<br />
Clayton, C. (1998). Elongation and clustering of glycosomes<br />
in Trypanosoma brucei overexpressing the<br />
glycosomal Pex11p. EMBO J. 17, 3542-3555.<br />
McAndrew, M., Graham, S. and Clayton, C.E. (1998).<br />
Testing promoter activity in the trypanosome genome:<br />
isolation of a metacyclic-type VSG promoter, and<br />
unexpected insights into RNA polymerase II transcription.<br />
Exp. Parasitol. 90, 65-76.<br />
Reviews<br />
Hotz, H.-R., Biebinger, S., Flaspohler, J., and Clayton,C.E.<br />
(1998). PARP Gene expression: regulation at<br />
many levels. Francqui Symposium Proceedings and<br />
Mol. Biochem. Parasitol. 91, 131-143.<br />
Clayton, C. E., et al. (1998). Genetic nomenclature for<br />
Trypanosoma and Leishmania. Mol. Biochem. Parasitol.<br />
97, 221-224.<br />
Clayton, C. E. (1999). Genetic manipulation of Kinetoplastida.<br />
Parasitol. Today 15, 372-378.<br />
Roditi, I., and Clayton, C. E. (1999). An unambiguous<br />
nomenclature for the major surface glycoprotein<br />
59
genes of the procyclic form of Trypanosoma brucei.<br />
Mol. Biochem. Parasit. 103, 99-100.<br />
Clayton, C.E., Maier, A., Lorenz, P., Blattner, J., Helfert,<br />
S., Krieger, S. (<strong>2000</strong>). Strange characteristics of<br />
kinetoplastid protists: targets for antiparasitic chemotherapy?<br />
Nova Acta Leopoldina 80, 15-25.<br />
Clayton, C.E. (1999). Why do we need standard<br />
genetic nomenclature for parasites? Acta Tropica (in<br />
press)<br />
THESES<br />
Diploma<br />
Ding, M. (1998). Charakterisierung von Organellen in<br />
Toxoplasma gondii.<br />
Herberger, M. (1999). Klonierung und Expression<br />
eines Cysteine Rich Acidic Integral Membrane<br />
(CRAM)-GFP-Fusionsgens in Trypanosoma brucei<br />
brucei.<br />
Dissertations<br />
Krieger, S. (1998). Die Trypanothionreduktase bei<br />
Trypanosoma brucei:Etablierung und Chatakterisie–<br />
rung einer TR-defizienten Zelllinie.<br />
Maier, A. (1999). Funktionelle Charakterisierung<br />
glykosomaler Membranproteine aus Trypanosoma<br />
brucei.<br />
60<br />
STRUCTURE OF THE GROUP<br />
E-mail: clayton@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong>: Clayton, Christine, Prof. Dr.<br />
Postdoctoral fellows<br />
Ansorge, Iris, Dr.<br />
Estévez, Antonio, PhD*<br />
Quilada, Luis, PhD*<br />
Voncken, Frank, PhD*<br />
Ph.D. students Ding, Martine, Dipl. Biol.*<br />
Drodzd, Maciej, Dipl. Biol.<br />
Guerra-Giraldez, Christina, MSc.<br />
Helfert, Sandra, Dipl. Biol.<br />
Irmer, Henriette, Dipl. Biol.<br />
Krieger, Stephan, Dipl. Biol.<br />
Maier, Alexan<strong>der</strong>, Dipl. Biol.<br />
Diploma students Ding, Martina *<br />
Schulreich, Sebastian *<br />
Herberger, Michael *<br />
Techn. assistant Hartmann, Claudia<br />
*part of the time reported<br />
Bernhard Dobberstein<br />
Protein Targeting and Intracellular<br />
Sorting<br />
Protein translocation across the membrane of the endoplasmic<br />
reticulum (ER) involves cytosolic chaperones,<br />
docking receptors, a translocation channel (translocon)<br />
and in some cases a “translocation motor” which<br />
drives the actual translocation (for review see Schatz<br />
and Dobberstein, 1996). Once in the ER, proteins are<br />
folded, modified and - after a quality control - packed<br />
into vesicles and transported to the Golgi complex and<br />
the trans-Golgi network. From there they can either<br />
be further transported to the plasma membrane or to<br />
organelles of the endosomal system. A major focus of<br />
the work of our group is the analysis of mechanisms<br />
involved in targeting proteins to the ER membrane<br />
and in their translocation across or insertion into this<br />
membrane. Special emphasis is on the control and<br />
regulation of these processes.<br />
Cotranslational targeting of nascent secretory and<br />
membrane proteins to the ER membrane involves<br />
the signal recognition particle (SRP) and its receptor<br />
(SRP-receptor or docking protein). SRP interacts with<br />
the signal sequence of the nascent proteins and mediates<br />
their transfer to the ER membrane. The SRP<br />
receptor catalyzes the release of the nascent chain<br />
from SRP and its insertion into the translocon. SRP<br />
comprizes a 7S RNA and six polypeptides of 9, 14, 19,<br />
54, 68 and 72 kDa. The SRP54 subunit is a GTPase<br />
to which the signal sequence of a nascent polypeptide<br />
chain binds. The SRP receptor consists of an α and β<br />
subunit both of which are GTPases. The translocation<br />
channel is formed by the Sec61p complex and several<br />
accessory proteins.<br />
Functional analysis of the three translocation-<br />
GTPases, SRP54, SRP receptor α and β<br />
G. Bacher and M. Pool<br />
The GTPase cycle of SRP54 was studied in its functional<br />
context, a ribosome nascent chain complex<br />
(RNC) and membranes containing different components<br />
of the translocation complex (Sec61p, TRAMp,<br />
SRP receptor). It was found that the ribosome stimulates<br />
GTP binding to SRP54 and that the GTP-bound<br />
state of SRP54 is important for high affinity binding<br />
of SRP to the SRP receptor α (Bacher et al. 1996).<br />
Two ribosomal proteins were identified that contact<br />
the GTPase domain and M-domain of SRP54 respectively<br />
(Pool and Dobberstein, in preparation). It is proposed<br />
that one of these proteins allows SRP to scan<br />
the RNC for the presence of a signal sequence and<br />
that the second protein functions in stimulating GDP /<br />
GTP exchange on SRP 54.<br />
The SRP receptor β subunit is anchored in the membrane<br />
and attaches SRP receptor α to the membrane.<br />
We have investigated GTP binding and hydrolysis of<br />
SRP receptor β. We found that SRP receptor β binds<br />
GTP with high affinity and interacts with ribosomes<br />
in the GTP-bound state. Subsequently, the ribosome<br />
increases the GTPase activity of SRP receptor β and<br />
thus functions as a GTPase activating component for<br />
SRP receptor β. We propose that SRP receptor β regulates<br />
the interaction of SRP receptor with the ribosome<br />
and thereby allows SRP receptor α to scan membrane<br />
bound ribosomes for the presence of SRP (Bacher,<br />
Pool and Dobberstein, 1999). Thus the two subunits<br />
of the SRP receptor regulate ribosome binding to the<br />
ER membrane and signal sequence insertion into the<br />
translocon.<br />
61
Protein Kinase C (PKC) phosphorylates essential<br />
translocon components<br />
O. Gruss with P. Feick, Homburg and R. Frank,<br />
<strong>ZMBH</strong> and S. Owen<br />
Secretion of proteins can occur in a constitutive or<br />
regulated manner. In the exocrine pancreas Ca ++ activated<br />
PKCs play a central role in the regulated secretion.<br />
In or<strong>der</strong> to identify possible targets for PKC<br />
at the ER membrane we have characterized PKCphosphorylated<br />
proteins of rough microsomes and the<br />
rough ER of intact cells. We found that essential components<br />
of the translocation machinery become phosphorylated<br />
in a Ca ++ -dependent manner. Among the<br />
proteins were the α subunit of the SRP receptor, the<br />
β subunit of the Sec61p complex and the TRAM protein.<br />
Isoform -specific antibodies revealed the presence<br />
of PKC α and β on rough ER membranes. Purified<br />
PKCs from rat brain phosphorylated the same<br />
translocon components as the endogenous Ca ++ -<br />
dependent kinases. Phosphorylation of translocon proteins<br />
was also observed in vivo and could be stimulated<br />
by phorbol esters. Phosphorylation of microsomal<br />
proteins by PKCs increased protein translocation<br />
efficiency in vitro. This suggests phosphorylation<br />
as a further level of regulation of protein translocation<br />
across the ER membrane (Gruss et al., 1999).<br />
Protein complexes of the translocation site<br />
L. Wang<br />
Several proteins or protein complexes function at<br />
different stages of the targeting or translocation process.<br />
Among them are the targeting components (SRP<br />
and SRP-receptor) the translocon (Sec61p complex),<br />
translocon associated components (TRAMp, RAMP4)<br />
oligosaccharyl transferase (OST) and signal peptidase<br />
complex. Depending on their functional state they<br />
62<br />
assemble transiently into supercomplexes. This is particularly<br />
evident for the translocon which can be unengaged,<br />
involved in cotranslational or posttranslational<br />
translocation or function in retrotranslocation. To analyse<br />
protein complexes of the rough ER we used mild<br />
solubilisation of ER membrane proteins, fractionation<br />
and blue native PAGE. Consistent with their multiple<br />
engagements we find targeting and translocation components<br />
in distinct oligomeric complexes (Wang and<br />
Dobberstein, 1999).<br />
Translocation-pausing mediated by RAMP4<br />
K. Schrö<strong>der</strong>, B. Martoglio, M. Hofmann in collaboration<br />
with E. Hartman, Göttingen, S. Prehn, Berlin and<br />
T. Rapoport, Boston<br />
Passage of nascent polypeptides across the membrane<br />
of the ER proceeds through a proteinaceous, aqueous<br />
channel formed by the Sec61p complex. To identify<br />
components that may regulate translocation by interacting<br />
with nascent polypeptides in the translocon,<br />
we used site-specific photo-crosslinking. Crosslinkers<br />
were placed around a consensus site for Asn-linked<br />
glycosylation. As a model protein we used the MHC<br />
class II-associated Invariant chain, which contains two<br />
closely spaced N-glycosylation sites. We found that a<br />
region C-terminal of the two consensus glycosylation<br />
sites in the Invariant chain binds in a hydrophobic<br />
interaction to RAMP4, a previously identified Ribosome<br />
Associated Membrane Protein (Görlich and<br />
Rapoport (1993) Cell, 75, 615-630). RAMP4 is a<br />
small, tail anchored protein of 66 amino acid residues.<br />
The interaction of RAMP4 with Ii occurred when<br />
nascent Ii chains reached a length of 170 amino<br />
acid residues and persisted until Ii chain completion,<br />
suggesting translocational pausing (Nakahara et al.<br />
(1994) J. Biol. Chem., 269, 7617-7622). Site-directed<br />
mutagenesis revealed that the region of Ii interacting<br />
with RAMP4 contains essential hydrophobic amino<br />
acid residues. Exchange of these residues for serines<br />
led to a reduced interaction with RAMP4 and inefficient<br />
N-glycosylation. We propose that RAMP4 controls<br />
modification of Ii and possibly also of other<br />
secretory and membrane proteins containing specific<br />
RAMP4-interacting sequences. Efficient or variable<br />
glycosylation of Ii may contribute to its capacity to<br />
modulate antigen presentation by MHC class II molecules<br />
(Schrö<strong>der</strong> et al., 1999).<br />
Figure 1: Hypothetical model of control of Ii chain glycosylation<br />
by RAMP4. Three stages of Ii insertion into the membrane<br />
are depicted. (I) Nascent Ii inserts into the translocon<br />
in a loop-like fashion. (II) When ~170 amino acid residues<br />
have been polymerized Ii interacts via its RIS (white box) with<br />
RAMP4. (III) Translocation is arrested and OST brought into<br />
position to efficiently glycosylate Ii. Upon chain completion, Ii<br />
detaches from RAMP4. Oligosaccharides are shown as forked<br />
structures.<br />
Signal sequences - more than just greasy peptides<br />
B. Martoglio, <strong>ZMBH</strong> / Zürich and M. Fröschke<br />
Signal sequences that target newly synthesized proteins<br />
to the ER contain a hydrophobic core region but<br />
otherwise show a great variation in both, overall length<br />
and amino acid sequence. Recently, it has become<br />
clear that this variation allows signal sequences to<br />
specify different modes of targeting and membrane<br />
insertion and even to perform functions after being<br />
cleaved from their parent protein. Signal sequences<br />
are therefore not simply greasy peptides but sophisticated,<br />
multipurpose peptides containing a wealth of<br />
functional information (Martoglio and Dobberstein,<br />
1998). Further work is directed to the functional analysis<br />
of unusually long signal sequences during targeting,<br />
proteolytic processing and after signal sequence<br />
cleavage.<br />
Signal sequences of the prion protein (PrP)<br />
C. Hölscher and U. C. Bach<br />
Prion protein (PrP) is synthesised at the membrane of<br />
the ER in three different topological forms, a secreted<br />
one (secPrP) and two that span the membrane in opposite<br />
orientations ( Ntm PrP and Ctm PrP). To identify signal<br />
sequences in PrP that determine the generation of the<br />
multiple forms of PrP we performed a deletion analysis.<br />
We found that PrP has – besides its N-terminal<br />
signal sequence- a C-terminal signal sequence that<br />
functions posttranslationally. The C-terminal signal<br />
sequence mainly mediates synthesis of Ctm PrP. Ctm PrP<br />
has been causally connected to certain inherited forms<br />
of neurodegeneration (Hegde et al. Nature, 402, 822 –<br />
826, 1999). We suggest that multiple forms of PrP are<br />
generated by the differential use of a N-terminal and<br />
C-terminal signal sequence (Hölscher, Bach and Dobberstein,<br />
in preparation).<br />
mRNA transport and protein localisation<br />
S. Frey and M. Seedorf in collaboration with R.<br />
Jansen, <strong>ZMBH</strong><br />
mRNA always exits the nucleus in a complex with<br />
RNA-binding proteins. Prior to translation complexes<br />
63
containing a subset of mRNAs are transported to<br />
specific regions inside the cell. This mechanism of<br />
local translation of transported mRNAs might play<br />
an important role to support protein targeting. The<br />
mRNA-binding protein vigilin displays unique features<br />
making it a good candidate to be a player in this<br />
process. Vigilin cofractionates with free and membrane<br />
bound ribosomes and shows an intracellular distribution<br />
similar as the endoplasmatic reticulum (ER).<br />
The protein consists almost exclusivly of RNA-binding<br />
domains. These domains provide multiple binding<br />
sites for messenger and ribosomal RNAs. To study the<br />
function of this unique RNA-binding protein we use<br />
budding yeast as a model system. Yeast expresses the<br />
structurally closely related protein Scp160p, which<br />
shows a similar intracellular distribution as vigilin.<br />
The SCP160 gene is not required for cell growth but<br />
a mutant without SCP160 fail to localize one specific<br />
mRNA. The majority of Scp160 protein associates<br />
with translating ribosomes, suggesting a function<br />
in the control of translation. Strong expression of<br />
Scp160p abolishes growth and reduces translation<br />
rates of a subset of so far unknown proteins. Cytoplasmic<br />
steady state localisation of Scp160p is independent<br />
of bulk mRNA export from the nucleus supporting<br />
the idea that Scp160p function is primary cytoplasmic<br />
and that the protein is not exported from the<br />
nucleus as constituent of a mRNA complex. Scp160p<br />
bears a masked nuclear import signal indicating that<br />
the function of Scp160p might be regulated by relocation<br />
of the protein to the nucleus.<br />
Microsomal ATP-Transport in yeast<br />
G. Konrad, T. Schlecker and P. Mayinger in collaboration<br />
with V. Bankaitis, Birmingham, Alabama<br />
ATP Transport into the ER is essential for ATP con-<br />
64<br />
suming reactions inside the ER lumen, including protein<br />
translocation and protein folding. We have characterized<br />
this nucleotide transport reaction as a specific<br />
ADP/ATP antiport and we have identified the<br />
Sac1 protein as an important regulator for this process.<br />
Sac1p is an ER and Golgi membrane protein with<br />
homology to a number of yeast and mammalian inositol<br />
phosphatases. It was shown recently that the N-terminal<br />
portion of Sac1p displays polyphosphoinositide<br />
phosphatase activity with unique enzymatic specificity.<br />
Consistent with these findings we obtained strong<br />
evidence in vitro that microsomal ATP transport is<br />
regulated via specific phosphoinositides.<br />
Cellular role of Sac1p in phosphoinositide<br />
signaling<br />
A. Then<br />
Sac1p plays additional roles in the regulation of<br />
intracellular membrane trafficking, phospholipid metabolism<br />
and the actin cytoskeleton. To elucidate the cellular<br />
function of Sac1p in more detail, we performed<br />
a synthetic lethal screen. In this screen we identified a<br />
novel allele of the SLT2 gene, which results in a severe<br />
growth defect when combined with a disruption of the<br />
SAC1 gene. The SLT2 gene codes for a PKC1-dependent<br />
MAP kinase, which plays an important role in<br />
cell wall integrity and in organization of cortical actin<br />
and performs an essential function in the context of<br />
a sac1D background. This genetic interaction with<br />
SLT2 defines a novel important function of the SAC1<br />
gene in regulating cell wall biogenesis and polarized<br />
secretion. Epistatic analysis places the SAC1 gene parallel<br />
to the PKC1 dependent SLT2 MAP kinase, linking<br />
this kinase cascade to phosphoinositide signaling.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(SFB 352 “<strong>Molekulare</strong> Mechanismen intrazellulärer<br />
Transportprozesse”, Graduiertenkolleg “<strong>Molekulare</strong><br />
Zellbiologie”), from the Landesforschungsschwerpunkt<br />
“Protein-Faltung und -Transport: Mechanismen<br />
und Pathobiochemie”, from the EU (“Signal Recognition<br />
Particle-Network”) and from the Fonds <strong>der</strong> Chemischen<br />
Industrie.<br />
PUBLICATIONS<br />
Martoglio, B., Hauser, S. and Dobberstein, B. (1998).<br />
Cotranslational translocation of proteins into microsomes<br />
<strong>der</strong>ived from the rough endoplasmic reticulum<br />
of mammalian cells. In Cell Biology, A Laboratory<br />
Handbook. Academic Press, 2nd ed. Vol. 2, pp.<br />
265-273.<br />
Graf, R., Brunner, J., Dobberstein, B. and Martoglio,<br />
B. (1998). Probing the molecular environment of proteins<br />
by site-specific photocrosslinking. In Cell Biology,<br />
A Laboratory Handbook. Academic Press 2nd ed.<br />
Vol 4, pp. 495-501.<br />
Martoglio, B. & Dobberstein, B. (1998). Signal<br />
sequences – more than just greasy peptides. Trends in<br />
Cell Biology 8, 410-415.<br />
Dube, P., Bacher, G., Stark, H., Müller, F., Zemlin, F.,<br />
van Heel, M. and Brimacombe, R. (1998). Correlation<br />
of the expansion segments in mammalian rRNA<br />
with the fine structure of the 80S ribosome; a cryoelectron<br />
microscopic reconstruction of the rabbit reticulocyte<br />
ribosome at 21A resolution. J. Mol. Biol. 279,<br />
403-421.<br />
Otter-Nilsson, M., Hendriks, R., Pecheur-Huet, E.I.,<br />
Hoekstra, D. and Nilsson, T. (1999). Cytosolic<br />
ATPases, p97 and NSF, are sufficient to mediate rapid<br />
membrane fusion. EMBO J. 18, 2074-2083.<br />
Gruss, O.J., Feick, P., Frank, R. and Dobberstein, B.<br />
(1999). Phosphorylation of components of the ER<br />
translocation site. Eur. J. Biochem. 260, 785-793.<br />
Schrö<strong>der</strong>, K., Martoglio, B., Hofmann, M., Hölscher,<br />
C., Hartmann, E., Prehn, S., Rapoport, T.A. and Dobberstein,<br />
B. (1999). Control of glycosylation of MHC<br />
class II-associated invariant chain by translocon-associated<br />
RAMP4. EMBO J. 18, 4804-4815.<br />
Bacher, G., Pool, M. and Dobberstein, B. (1999). The<br />
ribosome regulates the GTPase of the β-subunit of the<br />
signal recognition particle receptor. J. Cell Biol. 146,<br />
723-730.<br />
Wang, L. and Dobberstein, B. (1999). Oligomeric<br />
complexes involved in translocation of proteins across<br />
the membrane of the endoplasmic reticulum. FEBS<br />
Lett. 457, 316-22.<br />
Hofmann , M.W., Höning, S., Rodinov, D., Dobberstein,<br />
B., von Figura, K. and Bakke, O. (1999).<br />
The leucine-based sorting motifs in the cytoplasmic<br />
domain of the invariant chain are recognized by the<br />
medium chains of the clathrin adaptors AP1 and AP2.<br />
J. Biol. Chem. 274, 36153-36158.<br />
Kochendörfer, K.-U., Then, A. R., Kearns, B. G.,<br />
Bankaitis, V. A., and Mayinger, P. (1999). Sac1p plays<br />
a crucial role in microsomal ATP transport, which<br />
is distinct from its function in Golgi phospholipid<br />
metabolism. EMBO J. 18, 1506-1515.<br />
65
Then, A. R., Berger, J., Schwarz, H., and Mayinger, P.<br />
(<strong>2000</strong>). Sac1p regulates yeast cell wall organization in<br />
a signaling pathway that is functionally related to the<br />
Slt2p MAP kinase cascade. (in press).<br />
THESES<br />
Diploma<br />
Grimm, O. (1999). Isolation and partial characterization<br />
of proteins interacting with SRP-receptor.<br />
Schlecker, T. (1999). Einfluß <strong>der</strong> intrazellulären Lokalisation<br />
von Sac1p auf die Regulation des mikro–<br />
somalen ATP-Transports.<br />
Dissertations<br />
Gruß, O. (1998). Regulation <strong>der</strong> Translokation neusyn–<br />
thetisierter Polypeptide über die Membran des rauhen<br />
endoplasmatischen Retikulums.<br />
Then, A. (1999). Identifizierung von Faktoren die<br />
funktionell mit dem Sac1p Protein in Saccharomyces<br />
cerevisiae interagieren.<br />
STRUCTURE OF THE GROUP<br />
E-mail: dobberstein@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Dobberstein, Bernhard, Prof. Dr.<br />
Research<br />
associates Mayinger, Peter, Dr.<br />
Seedorf, Matthias, Dr.<br />
Postdoctoral<br />
fellows Barrett, Jeannie, Dr.*<br />
Hendriks, Robertus, Dr.*<br />
66<br />
Hölscher, Christina, Dr.<br />
Kipp, Bettina, Dr.*<br />
Martoglio, Bruno, Dr.*<br />
Owens, Sue, Dr.*<br />
Pool, Martin, Dr.<br />
Wang, Lin, Dr.*<br />
Ph.D. students Frey, Steffen,*<br />
Fröschke, Marc<br />
Gruß, Oliver*<br />
Konrad, Gerlinde*<br />
Then, Angela<br />
Diploma students Grimm, Oliver*<br />
Schlecker, Tanja*<br />
Techn. assistants Bach, Ute<br />
Meese, Klaus<br />
*part of the time reported<br />
Dirk Görlich<br />
Nucleocytoplasmic Transport<br />
The need for nuclear transport<br />
The nuclear envelope (NE) divides eukaryotic cells<br />
into a nuclear and a cytoplasmic compartment and<br />
thereby necessitates exchange between nucleus and<br />
cytoplasm. Not only must all nuclear proteins, such as<br />
histones and transcription factors, be imported from<br />
the cytoplasm, but also, tRNA, rRNA and mRNA are<br />
synthesised by transcription in the nucleus and need to<br />
be exported to the cytoplasm where they function in<br />
translation. Nuclear-cytoplasmic transport constitutes<br />
a tremendous activity that requires consi<strong>der</strong>able cellular<br />
resources and involves probably > 100 different<br />
and often very abundant proteins. However, these<br />
expenses pay off as indicated by the fact that only<br />
eukaryotes evolved into complex multicellular organisms.<br />
One can think of several reasons why a cell<br />
nucleus is required for such cellular complexity. First,<br />
the containment of the genome in a specialised organelle<br />
certainly improves genetic stability and allows<br />
eukaryotes to handle more genetic information than<br />
prokaryotes. Second, this compartmentation permits<br />
key cellular events to be regulated at a level that<br />
is unavailable to prokaryotes, e.g. by controlling the<br />
access of transcriptional regulators to chromatin. A<br />
further reason is the composition of typical eukaryotic<br />
genes from exons and introns, which requires primary<br />
transcripts to be spliced before translation may occur.<br />
Translation of unspliced pre-mRNAs would produce<br />
proteins that are not only non-functional but which<br />
may potentially act as dominant-negative inhibitors.<br />
The confinement of transcription and translation to<br />
distinct compartments can therefore be consi<strong>der</strong>ed a<br />
good solution to avoid this problem.<br />
Importin beta-like transport receptors<br />
P. Schwarzmaier, U. Kutay, G. Lipowsky, F. Paraskeva,<br />
S. Jäckel, K. Ribbeck, K. Dean, in collaboration<br />
with E. Hartmann (Univ. Göttingen), R. Kraft and<br />
S. Kostka (MDC, Berlin)<br />
Nuclear-cytoplasmic transport proceeds through<br />
nuclear pore complexes (NPCs) which allow passive<br />
diffusion of small molecules up to 20-40kD and also<br />
active, i.e., carrier-mediated transport of macromolecules.<br />
This active transport can occur along a great<br />
variety of distinct pathways, many of which are mediated<br />
by transport receptors that are at least distantly<br />
related to importin β. Transport receptors that confer<br />
import into the nucleus are referred to as importins,<br />
while export mediators are called exportins.<br />
A major objective of our laboratory has been to obtain<br />
a detailed and comprehensive overview of the various<br />
nuclear transport pathways. To identify novel transport<br />
receptors, we have employed a biochemical approach<br />
combined with data base searches and have so far<br />
arrived at 22 mammalian members of the importin β<br />
superfamily. In the following, we will briefly summarise<br />
some of our recent insights in the mechanistic<br />
principles of transport receptor function and elaborate<br />
some nuclear transport pathways in more detail.<br />
Mechanistic principles of transport receptor<br />
function<br />
Importins or exportins are capable of mediating multiple<br />
rounds of transport, which requires them to shuttle<br />
continuously between nucleus and cytoplasm. A given<br />
transport cycle, however, can only be productive if<br />
the corresponding transport receptor carries cargo in<br />
one direction only and returns „empty“ to the original<br />
compartment. This asymmetry in the transport cycles<br />
can be explained by the RanGTP-gradient model. Ran<br />
67
switches between a GDP- and a GTP-bound form. Its<br />
nucleotide-bound state is controlled by a number of<br />
activities whose asymmetric nucleocytoplasmic distribution<br />
is thought to result in a high nuclear RanGTP<br />
concentration and very low RanGTP levels in the cytoplasm.<br />
Importin-β like transport receptors are RanGTP<br />
binding proteins that respond to this gradient by<br />
loading and unloading their cargo in the appropriate<br />
compartment. Importins bind their substrates in the<br />
absence of RanGTP, i.e. in the cytoplasm and release<br />
them upon encountering RanGTP in the nucleus. The<br />
importins then return to the cytoplasm where RanGTP<br />
is released (involving GTP hydrolysis), thereby allowing<br />
the importin to bind and import the next cargo molecule.<br />
Exportins respond to the RanGTP gradient in<br />
exactly the opposite way; they bind their cargo preferentially<br />
in the presence of RanGTP in the nucleus,<br />
where they form a trimeric substrate-exportin-RanGTP<br />
complex. The trimeric export complex is disassembled<br />
after export to the cytoplasm and the „empty“ exportin<br />
can re-enter the nucleus to participate in another round<br />
of export.<br />
Transport receptors bind their transport substrates in<br />
many cases directly, an example being nuclear export<br />
of tRNA by exportin-t (see below). In other cases, substrate<br />
recognition is more complicated and involves an<br />
adapter molecule. The best studied example for that is<br />
the classical NLS import pathway, where the NLS is<br />
recognised by the adapter importin α, which in turn<br />
binds the actual transport receptor importin beta. This<br />
complicates the corresponding transport cycle consi<strong>der</strong>ably<br />
in that not only importin β, but also importin<br />
α needs to be return to the cytoplasm. Interestingly,<br />
importin α employs a specialised exportin, namely<br />
CAS, for this purpose.<br />
68<br />
Figure 1: Transport cycles of Importins (Imp) and exportins<br />
(Exp). For details, please see main text.<br />
Recycling of snurportin 1 back to the cytoplasm<br />
F. Paraskeva, U. Kutay, in collaboration with R. Lührmann<br />
(Univ. Marburg), F.R. Bischoff (DKFZ, Heidelberg),<br />
E. Izaurralde (EMBL, Heidelberg)<br />
Snurportin 1 functions as an import adapter for U<br />
snRNPs. It recognises the m 3 G-cap import signal of<br />
U snRNPs and also binds Importin β which in turn<br />
is the actual mediator of import. Just as importin α,<br />
also snurportin 1 needs to be returned to the cytoplasm<br />
after each round of import in or<strong>der</strong> to accomplish further<br />
import cycles. We found that this recycling is<br />
mediated by CRM1. CRM1 was originally identified<br />
as the exportin specific for leucine-rich nuclear export<br />
signals (NESs). However, the CRM1/ snurportin 1<br />
interaction differs significantly from that with other<br />
export substrates: Firstly, snurportin 1 binds CRM1<br />
not through a short, leucine-rich signal, but instead<br />
through a large domain that comprises more than 200<br />
residues. Secondly, snurportin 1 binds CRM1 100<br />
times more tightly than an NES from the HIV Rev<br />
protein. In addition, snurportin 1 can bind either the<br />
m 3 G-cap import signal or CRM1, but not both at the<br />
same time. This property ensures that CRM1 exports<br />
only those snurportin 1 molecules which have already<br />
released their cargo and thereby allows snurportin 1 to<br />
mediate productive import cycles.<br />
tRNA export<br />
U. Kutay, G. Liposwsky, P. Schwarzmaier, in collaboration<br />
with E. Izaurralde (EMBL, Heidelberg) and<br />
F.R. Bischoff (DKFZ, Heidelberg)<br />
tRNAs are synthesised as precursor molecules (pretRNAs),<br />
mature to functional tRNA and finally get<br />
exported to the cytoplasm. There, they participate in<br />
cycles of aminoacylation, binding to the elongation<br />
factor eE1A and function in translation. We have identified<br />
exportin-t as the tRNA-specific exportin and<br />
showed that it functions according to the exportinparadigm<br />
described above. The maturation of pretRNAs<br />
to functional tRNAs involves trimming of the<br />
3´and 5´ends, post-transcriptional addition of the 3´<br />
CCA end to which the amino acid is later attached,<br />
and the modification of a number of nucleosides. It<br />
is quite remarkable that exportin-t preferentially binds<br />
and exports mature tRNA which contain correctly processed<br />
3´and 5´ends and the appropriate nucleoside<br />
modifications. Exportin-t mediated export thus constitutes<br />
a proof-reading or quality-control mechanism<br />
that co-ordinates RNA processing with export and<br />
thereby helps to ensure that only functional tRNA<br />
arrives in the cytoplasm.<br />
Nuclear import of ribosomal proteins and<br />
histone H1<br />
S. Jäckel<br />
The biogenesis of ribosomes is a very complex process<br />
that also involves nuclear import and export events:<br />
Ribosomal proteins are first imported from the cytoplasm,<br />
assemble with rRNA in the nucleolus to form<br />
ribosomal subunits which are then finally re-exported<br />
to the cytoplasm.<br />
We have studied nuclear import of ribosomal proteins<br />
in higher eukaryotes, focusing on rpL23a. We found<br />
that at least 4 distinct transport receptors, namely<br />
importin β, transportin, importin 5 and importin 7<br />
can directly bind and import rpL23a. This not only<br />
assigned the first functions to importin 5 and 7,<br />
but showed an apparently quite common principle<br />
in nuclear import, namely that some substrates can<br />
„choose“ between several carriers. L23a binds through<br />
the same domain or „import signal“ to each of the 4<br />
transport receptors.<br />
69
Ribosomal proteins are generally very basic and have<br />
a high tendency to precipitate and aggregate before<br />
their assembly into ribosomal subunits. It is therefore<br />
important to note that the importins shield rather large<br />
domains of the ribosomal proteins, thereby preventing<br />
undesired interactions in the cytoplasm. Thus, the<br />
importins not only fulfil an import function but also<br />
act as chaperonins.<br />
Histones are also very basic and cause similar aggregation<br />
problems as ribosomal proteins. This posed the<br />
questions of whether histones are imported the same<br />
way as ribosomal proteins. Surprisingly, we found that<br />
the histone H1 cannot be imported into the nucleus<br />
by any „single“ importin. Instead, the active species<br />
in H1 import is an importin β/importin 7 heterodimer.<br />
Both importins appear to contact the histone, probably<br />
because a single import receptor is not sufficient<br />
to completely wrap the extended and extremely basic<br />
domain of this cargo.<br />
Recycling of Ran back to the nucleus<br />
K. Ribbeck, G. Lipowsky<br />
A key element of current models for importin and<br />
exportin function is that these factors normally enter<br />
the nucleus Ran-free and exit as a complex with<br />
RanGTP. Importins carry cargo in their Ran-free form,<br />
while exportins do so when complexed with RanGTP.<br />
Both have in common to constantly deplete Ran from<br />
the nucleus (see Fig. 1). These transport cycles can<br />
then only be maintained by an extremely efficient<br />
nuclear import of Ran. The assumption that each<br />
transport cycle of an importin β-like transport receptor<br />
results in export of one Ran molecule would imply<br />
that Ran crosses the nuclear envelope as frequently<br />
as all these receptors together. For reasons of stoichiometry<br />
it then appears impossible that Ran itself<br />
70<br />
is imported in a conventional way by an importin β<br />
family transport receptor.<br />
We have established an in vitro system to study nuclear<br />
import of Ran and found that this process requires<br />
some soluble nuclear transport factor. Another key<br />
observation was that wild type Ran was efficiently<br />
imported, but the RanQ69L mutant was not. We could<br />
thus use immobilised wild type Ran to deplete its<br />
import factor from the cytosol, while depletion with<br />
immobilised RanQ69L had no effect. Examination of<br />
the eluates revealed that a 15 kD protein had bound to<br />
immobilised wild type Ran, but was absent from the<br />
RanQ69-bound fraction. The 15 kD protein was identified<br />
as nuclear transport factor 2 (NTF2) and indeed,<br />
recombinant NTF2 could restore the Ran-import activity<br />
of the depleted cytosol (see Fig. 2). This established<br />
that Ran uses for its own import a unique import<br />
pathway that is mediated by NTF2.<br />
The Ran-Transport cycles can thus be described as<br />
follows (see Fig. 1): RanGDP is bound by NTF2 in<br />
the cytoplasm and translocated into the nucleus. The<br />
Figure 2: Effect of NTF2 on nuclear import of Ran. Figure<br />
shows import of fluorescent Ran into nuclei of permeabilised<br />
cells, performed either in the absence or presence of<br />
NTF2.<br />
RanGEF (nucleotide exchange factor) is a nuclear,<br />
chromatin-bound protein and converts RanGDP to<br />
RanGTP. RanGTP can now bind to an importin and<br />
displace the import cargo or it can assemble into<br />
a trimeric cargo/ exportin/ RanGTP complex. The<br />
RanGTP/ transport receptor complexes are then transferred<br />
to the cytoplasm, where they become disassembled<br />
by the concerted action of RanBP1 and<br />
the RanGTPase activating protein RanGAP (both of<br />
which are excluded from the nuclei). This disassembly<br />
results in the hydrolysis of the Ran-bound GTP molecule,<br />
cargo release from exportins and it allows importins<br />
to bind another substrate molecule. RanGDP can<br />
now enter into another transport cycle.<br />
Mechanism and energetics of NPC-passage<br />
K. Ribbeck<br />
The nuclear transport system is capable of accumulating<br />
cargo molecules against a gradient of chemical<br />
activity, which is obviously an energy-consuming task.<br />
The question of how the input of metabolic energy<br />
is coupled to active transport across the nuclear envelope<br />
has been a central problem in nucleocytoplasmic<br />
transport. As indicated in the scheme of Fig. 1, each<br />
importin- or exportin-dependent transport cycle results<br />
in the hydrolysis of one molecule of GTP. This happens<br />
when RanGTP/transport receptor complexes are<br />
disassembled in the cytoplasm. A surprising recent<br />
finding by us and others has been that this single GTPhydrolysis<br />
event constitutes the sole input of energy<br />
into these transport cycles and that the actual translocation<br />
process is not directly coupled to any nucleotide<br />
hydrolysis or any other irreversible step. This<br />
meant a radical change in concept as the actual translocation<br />
through NPCs had previously been assumed<br />
to be driven by NTP-hydrolysis.<br />
There is an interesting parallel between the energetics<br />
of the just mentioned nucleocytoplasmic transport<br />
cycles and membrane-bound transporters for lowmolecular<br />
weight substances. These transporters can<br />
often use the chemical potential of a primary gradient<br />
(e.g. of Na + ions or protons) to accumulate another<br />
molecule against a gradient of chemical activity. One<br />
textbook example is uptake of amino acids by mammalian<br />
cells which is powered by the Na + ion gradient<br />
across the plasma membrane. The Na + ion concentration<br />
is lower inside the cell than outside and the transporter<br />
couples the uptake of an amino acid with an<br />
influx of Na+ ions, i.e. it symports the two molecules.<br />
Transport cycles of exportins and importins can be<br />
seen in a similar way in that they also use the chemical<br />
potential of a primary gradient, namely that of<br />
RanGTP, to drive the directed transport of cargo molecules.<br />
Exportins are symporters for export cargo<br />
and RanGTP (see Fig. 1). Conversely, importins can<br />
be consi<strong>der</strong>ed as antiporters that accomplish nuclear<br />
cargo accumulation against a gradient of chemical<br />
activity by „paying“ with the export of RanGTP that<br />
occurs down a gradient. The other constituents of the<br />
RanGTP system function to replenish the Ran gradient<br />
and use GTP-hydrolysis by Ran as the energysource<br />
for this purpose.<br />
The actual translocation through NPCs constitutes a<br />
fully reversible process that can be described as some<br />
kind of facilitated diffusion. The mechanistic basis for<br />
this process, however, is still unclear and for example,<br />
we do not yet un<strong>der</strong>stand why a cargo-transport receptor<br />
complex can pass the NPC so much faster than<br />
the cargo molecule alone. The mechanistics of NPCpassage<br />
will therefore be a major focus of our future<br />
research activity.<br />
71
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(SFB 352 “<strong>Molekulare</strong> Mechanismen intrazellulärer<br />
Transportprozesse”, DFG-Schwerpunkt “Funktionelle<br />
Architektur des Zellkerns”, Graduiertenkolleg<br />
“<strong>Molekulare</strong> Zellbiologie”), from the Human Frontier<br />
Science Programme Organization and from the Fonds<br />
<strong>der</strong> Chemischen Industrie.<br />
PUBLICATIONS<br />
Kutay, U., Lipowski, G., Izaurralde, E., Bischoff,<br />
F.R., Schwarzmaier, P, Hartmann, E., and Görlich,<br />
D. (1998). Identification of a tRNA-specific nuclear<br />
export receptor. Mol. Cell. 1, 359-369.<br />
Görlich, D. (1998). Transport into and out of the cell<br />
nucleus (review). EMBO J. 17, 2721-2727.<br />
Jäkel, S., and Görlich, D. (1998). Importin β, transportin,<br />
RanBP5 and RanBP7 mediate nuclear import of<br />
ribosomal proteins in mammalian cells. EMBO J. 17,<br />
4491-4502.<br />
Ribbeck, K., Lipowsky, G., Kent, H.M., Stewart, M.,<br />
and Görlich, D. (1998). NTF2 mediates nuclear import<br />
of Ran. EMBO J. 17, 6587-6598.<br />
Ribbeck, K., Kutay, U., Paraskeva, E. and Görlich, D.<br />
(1999). The translocation of transportin-cargo complexes<br />
through nuclear pores is independent of Ran<br />
and energy. Curr. Biol. 9, 47-50.<br />
Lipowsky, G., Bischoff, F.R., Izaurralde, E., Kutay,<br />
U., Schäfer, S. Gross, H.J., Hildburg Beier, H. and<br />
Görlich, D. (1999). Coordination of tRNA nuclear<br />
72<br />
export with processing of tRNA. RNA 5, 539-549.<br />
Ziemienowicz, A., Görlich, D., Lanka, E., Hohn, B.,<br />
and Rossi, L. (1999). Import of DNA into mammalian<br />
nuclei by proteins originating from a plant pathogenic<br />
bacterium. Proc. Natl. Acad. Sci. USA 96, 3729-33.<br />
Jäkel, S., Albig, W., Schwamborn, K., Doenecke, D.,<br />
and Görlich, D. (1999). The importinβ/ importin7 heterodimer<br />
is a functional nuclear import receptor for<br />
histone H1. EMBO J. 18, 2411-2423.<br />
Paraskeva, E., Izaurralde, E., Bischoff, F.R., Huber, J.,<br />
Kutay, U., Hartmann, E., Lührmann, R., and Görlich,<br />
D. (1999). Crm1p-mediated recycling of snurportin 1<br />
to the cytoplasm. J. Cell Biol. 145, 255-264.<br />
Görlich, D. and Kutay, U. (1999). Transport between<br />
the cell nucleus and the cytoplasm. Annu. Rev. Cell<br />
Dev. Biol. 15, 607-660.<br />
Vetter, I.R., Arndt, A. Kutay, U., Görlich, D. and<br />
Wittinghofer, A (1999). Structural view of the Ran-<br />
Importin β interaction at 2.3 Å resolution. Cell 97,<br />
635-646.<br />
Jullien, D., Görlich, D., Laemmli, U.K., and Adachi,<br />
Y (1999). Nuclear import of RPA in Xenopus egg<br />
extracts requires a novel protein XRIPα but not importin<br />
α. EMBO J. 18, 4348-4358.<br />
Bayliss, R., Ribbeck, K., Akin, D., Kent, H.M., Feldherr,<br />
C.M., Görlich, D., and Stewart, M. (1999).<br />
Interaction between NTF2 and xFxFG-containing<br />
nucleoporins is required to mediate nuclear import of<br />
RanGDP. J. Mol. Biol. 293, 579-93.<br />
Köhler, M., Speck, C., Christiansen, M., Bischoff,<br />
F.R., Prehn, S., Haller, H., Görlich, D., and Hartmann,<br />
E. (1999). Evidence for distinct substrate specificities<br />
of importin alpha family members in nuclear protein<br />
import. Mol. Cell. Biol. 19, 7782-91.<br />
Bachi, A, Braun, I.C., Rodrigues, J.P., Pante, N.,<br />
Ribbeck, K., von Kobbe, C., Kutay, U., Wilm, M.,<br />
Görlich, D., Carmo-Fonseca, M., and Izaurralde, E.<br />
(<strong>2000</strong>). The C-terminal domain of TAP interacts with<br />
the nuclear pore complex and promotes export of<br />
specific CTE-bearing RNA substrates. RNA J., (in<br />
press).<br />
Bannister, A.J., Miska, E.A., Görlich, D. and Kouzarides,<br />
T. (<strong>2000</strong>). Acetylation of importin α nuclear<br />
import factors by CBP/p300. Curr. Biol. 10, 467-470.<br />
Lipowsky, G., Bischoff, F.R., Schwarzmaier, P., Kraft,<br />
R., Hartmann, E., Kutay, U., and Görlich, D. (<strong>2000</strong>).<br />
Exportin 4: a mediator of a novel nuclear export pathway<br />
in higher eukaryotes. EMBO J., (in press).<br />
STRUCTURE OF THE GROUP<br />
E-mail: dg@zmbh.uni-heidelberg.de (Dirk Görlich)<br />
Group lea<strong>der</strong> Görlich, Dirk, Dr.<br />
Postdoctoral<br />
fellows Kutay, Ulrike, Dr. *<br />
Paraskeva, Froso, Dr.<br />
Dean, Kelly, Dr.*<br />
Mingot, José-Manuel, Dr. *<br />
Ph.D. students Lipowsky, Gerd, Dipl. Biol.<br />
Jäckel, Stefan, Dipl. Biol.<br />
Ribbeck, Katharina, Dipl. Biol.<br />
Technical assistant Schwarzmaier, Petra<br />
* part of the time reported<br />
73
Richard Herrmann<br />
Molecular Biology of the Bacterium<br />
Mycoplasma pneumoniae<br />
So far, the genomes of 26 different prokaryotes have<br />
been completely sequenced and published. Among<br />
those are the prototype of the gram-positive bacteria,<br />
Bacillus subtilis, and of the gram-negative bacteria,<br />
Escherichia coli, and several archaea from extreme<br />
habitats. In addition, the first genomes of two eukaryotes,<br />
baker‘s yeast (Saccharomyces cerevisiae) and<br />
the nematode Caenorhabditis elegans, are available.<br />
We expect, by the year <strong>2000</strong> the genome of the first<br />
plant, Arabidopsis thaliana, and of the fruit fly Drosophila<br />
melanogaster will be also sequenced. The annotation<br />
of these genome sequences has made it possible<br />
to predict functions for at least half of the proposed<br />
genes and permits an un<strong>der</strong>standing of the metabolic<br />
pathways and the synthesis of macromolecules. But<br />
the sequence and annotation are not sufficient to<br />
un<strong>der</strong>stand the two key processes characterizing all<br />
living systems that are metabolism, defined as the sum<br />
of all chemical reactions taking place in a cell and<br />
reproduction. Nevertheless, a genome sequence and<br />
annotation lead us directly to several experimental<br />
approaches that can bring us closer to an un<strong>der</strong>standing<br />
of the total biology of a living cell.<br />
Such experiments will include:<br />
• Identification of all genes and their functional<br />
assignment,<br />
• Classification of essential and non-essential genes<br />
(C. Hutchison et al., 1999),<br />
• Transcription and translation profiles,<br />
• Analysis of quantitative changes at the level of<br />
gene expression depending on growth conditions,<br />
e.g. growth with or without a host cell,<br />
• Analysis of the mechanisms of gene regulation,<br />
• Studies on the interaction between gene products.<br />
74<br />
Despite the progress in methodology, such analyses<br />
are still very complex and, therefore, we profess they<br />
can be done most readily in a simple cell with a small<br />
number of gene products. We have chosen the human<br />
pathogenic bacterium Mycoplasma pneumoniae as a<br />
model organism to study the biochemistry and molecular<br />
biology of a minimal cell.<br />
Mycoplasma pneumoniae is one of the smallest bacteria<br />
known with a genome size of 816 kbp and a cell<br />
diameter of about 0.5µ. It colonizes as a human pathogen<br />
the respiratory tract causing an atypical pneumonia.<br />
M. pneumoniae is host dependent in nature, but it<br />
can be grown in the laboratory without a host cell in<br />
a rich medium containing 5-20% serum. The cholesterol<br />
in the serum is essential for growth and is incorporated<br />
into the cell membrane. Lacking a cell wall<br />
the membrane of M. pneumoniae is the only outer<br />
layer that separates it from the environment.<br />
I. Gene expression and functional analysis<br />
Most of the functional assignments of proposed genes<br />
from genome sequencing projects are based on significant<br />
sequence similarities to genes/proteins with<br />
known function from other organisms. Besides the<br />
search for sequence similarities other more general<br />
approaches for functional assignments are possible.<br />
Our approach for a functional analysis combines gene<br />
expression studies with functional characterization.<br />
The assumption is that un<strong>der</strong> changing growth conditions<br />
individual genes might be differently expressed<br />
and that specific response to a certain stimulus like<br />
change in temperature, ionic strength or oxygen concentration,<br />
etc., will provide indications for function.<br />
Our gene expression studies of M. pneumoniae include<br />
all the proposed genes and they are being done at the<br />
level of transcription and translation. The first step in<br />
both analyses will be the establishment of a transcription<br />
(transcriptome) and translation (proteome) profile<br />
of M. pneumoniae grown un<strong>der</strong> standard conditions.<br />
These standard profiles will serve as references for<br />
profiles, which were generated from M. pneumoniae<br />
grown un<strong>der</strong> modified conditions.<br />
II. Transcription analysis<br />
J. Weiner, H. Göhlmann, C.U. Zimmermann,<br />
S. Schulz, E. Pirkl<br />
Our approach to transcription analyses was two-fold.<br />
First, the structure of mycoplasma promoters was<br />
studied and second, transcription of all proposed open<br />
reading frames (transcriptome) was monitored.<br />
Presently, little is un<strong>der</strong>stood of the structure of mycoplasma<br />
promoters. Since we are interested in un<strong>der</strong>standing<br />
regulation of gene expression of an organism<br />
possessing only one recognized sigma factor, promoter<br />
structures of M. pneumoniae had to be defined.<br />
For this reason the transcriptional start points of 22<br />
genes, which were transcribed at different rates (see<br />
below), were identified by primer extension analysis<br />
and the regions upstream and downstream were compared.<br />
Several possible –10 region promoter sequences<br />
were found, but there were no obvious conserved –35<br />
regions. Another unusual feature was the lack of an<br />
untranslated lea<strong>der</strong> region in front of the first start<br />
codon which could contain a ribosomal binding site.<br />
Search for other translation signals like a downstream<br />
box was also unsuccessful. Inspection of the region<br />
about 50 bases upstream of the proposed start codon<br />
for all other open reading frames revealed that in the<br />
majority of cases promoters with a –10 region but not<br />
with a –35 region were conserved.<br />
The first step of the transcriptome analysis is the<br />
establishment of a reference transcription profile for<br />
all proposed ORFs of M. pneumoniae. For this purpose,<br />
total RNA from bacteria grown un<strong>der</strong> standard<br />
laboratory conditions was isolated, reverse transcribed<br />
into single stranded complementary DNA and labeled<br />
with 33 P. Further, specific probes for all proposed<br />
ORFs were synthesized by polymerase chain reaction,<br />
immobilized on nylon membranes and tested<br />
for cross-hybridization with the labeled cDNA. About<br />
500 transcripts from different ORFs were detected.<br />
They could be divided into several classes according<br />
to signal strength that reflects the number of individual<br />
RNA species. We have no experimental evidence<br />
describing how transcription is regulated. However,<br />
we expect some information about the expression of<br />
individual genes, operons or regulatory networks from<br />
comparative analyses of transcription profiles from<br />
cells grown un<strong>der</strong> modified conditions, e.g., un<strong>der</strong> different<br />
temperatures (32°C, 37°C, 43°C), osmotic or<br />
oxygen stress etc. We are extending the transcriptome<br />
analysis to the intergenic regions. Among others, we<br />
found a highly abundant RNA species that has the<br />
potential to code for a 29 amino acid long cysteine<br />
rich protein. Experiments are in progress to identify<br />
this peptide in bacterial extracts.<br />
III. Proteome analysis<br />
J. Regula, B. Ueberle<br />
Proteome analysis involves the two-dimensional (2-D)<br />
gel electrophoresis for the separation of proteins<br />
according to isoelectric point (pI) and molecular mass,<br />
the knowledge of the sequences of all proteins of a<br />
cell and mass spectrometry for the characterization<br />
75
of individual proteins. The combination of peptide<br />
mass finger-printing and peptide fragmentation which<br />
matches the masses of in gel proteolytically generated<br />
peptides against theoretically digested proteins from<br />
the M. pneumoniae database, has proved to be very<br />
effective and reliable. Proteome analysis is limited by<br />
the sensitivity of mass spectrometry and the pH range<br />
of the first dimension of the two-dimensional gel electrophoresis.<br />
Presently proteins with isoelectric points<br />
Figure 1: 2-D-gel of a total protein extract from M. pneumoniae.<br />
First dimension (1-D) immobilized pH gradient,<br />
second dimension (2-D) 12,5% SDS-polyacrylamide gel.<br />
All stained protein spots (≈ 200) were assigned to the corresponding<br />
genes, but only the strongest spots are marked<br />
with a gene number.<br />
76<br />
Figure 2: Mock 2-D-gel of all proposed proteins of M.<br />
pneumoniae. Based on the predicted molecular mass and<br />
isoelectric point, each protein could be localized in this<br />
mock 2-D-gel. The yellow and blue dots represent the sum<br />
of all proposed proteins; the „blue“ proteins have been<br />
assigned to a gene.<br />
between 3-10 can only be separated routinely by<br />
immobilized pH gradients (Fig. 1), but the annotation<br />
of the genome sequence of M. pneumoniae predicts<br />
236 proteins with pIs greater than 10 (Fig. 2). When<br />
we analyze the protein extract of entire M. pneumoniae<br />
cells by two-dimensional gel electrophoresis<br />
and visualize the proteins by silver staining, about 450<br />
proteins can be detected and about 200 proteins by<br />
staining with the less sensitive Coomassie blue (Fig.<br />
1). So far, 200 proteins have been identified by mass<br />
spectrometry and assigned to the corresponding genes.<br />
We found in most cases a good correlation between<br />
DNA sequence based prediction of pI and mass of a<br />
protein and its final position within a two-dimensional<br />
gel. However, exceptions have been found where the<br />
experimentally <strong>der</strong>ived values were different. We suggest<br />
that in many of these exceptions posttranslational<br />
modification might have occurred. Furthermore,<br />
a comparison of transcriptome and proteome analysis<br />
shows that a relatively high copy number of a given<br />
transcript does not always correlate with a strong pro-<br />
tein signal and vice versa. Those genes could be interesting<br />
examples for regulation of gene expression at<br />
different levels. The proteome analysis is being done<br />
in cooperation with R. Frank, Heidelberg and A. Görg,<br />
München.<br />
IV. Is there a cytoskeleton-like structure in M.<br />
pneumoniae?<br />
J. Regula, A. Boonmee, W. Schaller<br />
The M. pneumoniae genome does not encode for a<br />
single gene known to be involved in the synthesis<br />
of a bacterial cell wall. M. pneumoniae is only surrounded<br />
by a cytoplasmic membrane that, exceptionally<br />
among bacteria, contains cholesterol as an essential<br />
component. Over the past twenty years evidence<br />
has accumulated that M. pneumoniae possesses a cytoskeleton-like<br />
structure, probably as a substitute for the<br />
missing cell wall. In analogy to the cytoskeleton of<br />
eukaryotic cells, such a structure could provide the<br />
necessary framework for maintaining and stabilizing<br />
the shape of M. pneumoniae, for motility and for the<br />
formation of an asymmetric cell.<br />
The first experimental evidence for a cytoskeleton-like<br />
structure in M. pneumoniae was provided by Meng<br />
and Pfister (1980) who detected fibrous structures<br />
by electron microscopy after treating M. pneumoniae<br />
cells with the nonionic detergent Triton X-100, which<br />
removed the membrane and the cytoplasm. These<br />
observations were confirmed and extended by several<br />
other researchers. These experiments and studies on<br />
the architecture and composition of eucaryotic cytoskeletons<br />
from cells which have been treated with the<br />
detergent Triton X-100 suggested that a cytoskeletonlike<br />
structure would also be enriched in the Triton<br />
X-100 insoluble fraction of M. pneumoniae.<br />
Therefore, we decided to determine the protein com-<br />
position of the Triton X-100 insoluble fraction of<br />
M. pneumoniae by 2-D-gel electrophoresis and mass<br />
spectrometry.<br />
Silver staining of 2-D gels of the Triton X-100 insoluble<br />
fraction revealed about 100 protein spots. By<br />
staining with colloidal Coomassie blue about 50 protein<br />
spots were visualized of which 41 were identified<br />
by determining the mass and the partial sequence of<br />
their tryptic peptides following enzymatic digestion.<br />
The identified proteins belonged to several functional<br />
categories, mainly energy metabolism, translation and<br />
heat shock response. In addition, we found lipoproteins<br />
and most of the proteins involved in cytadherence<br />
which were known to be components of the<br />
Triton X-100 insoluble fraction based on evidence<br />
from previous experiments. There were also 11 functionally<br />
unassigned proteins. The quantitatively most<br />
prevalent proteins were the heat shock protein DnaK,<br />
the elongation factor Tu and the subunits α and ß<br />
of the pyruvate dehydrogenase (PdhA, PdhB). To<br />
prove whether a cytoskeleton-like structure exists<br />
in Mycoplasma pneumoniae, further experiments are<br />
required.<br />
One of the promising newer methods to identify in<br />
vivo interacting proteins is the two-hybrid system in<br />
Saccharomyces cerevisiae. We started in cooperation<br />
with M. Kögel (LION Bioscience, Heidelberg) a twohybrid<br />
analysis with the aim of finding which of the<br />
proposed proteins of the cytoskeleton-like structure<br />
interact directly. We expect that this approach will<br />
also reveal new candidates for structural components.<br />
As starting material for our analysis we used the gene<br />
hmw2. Our selection is based on data from various<br />
laboratories indicating that the gene hmw2 codes for<br />
one of the key components in the formation of the<br />
cytoskeleton-like structures. So far, we have identified<br />
several interacting proteins. These results are now<br />
being verified by a second independent method.<br />
77
V. Genetic stability of infectious M. pneumoniae<br />
isolates<br />
W. Reiser, I. Catrein, M. Götzmann, W. Schaller,<br />
E. Pirkl<br />
M. pneumoniae causes sporadic endemic disease but<br />
epidemics have been reported to appear in certain<br />
time intervals. Infectious M. pneumoniae isolates from<br />
patients showed that either one of the two subtypes<br />
(I and II) was the causative agent in an outbreak.<br />
The prototypes of these subtypes are M. pneumoniae<br />
M129(I) and M. pneumonia FH(II). The basis for their<br />
classification is the P1 adhesin, which is essential for<br />
adherence of the bacterium to its host. To obtain more<br />
information about the differences between the two<br />
prototypes and various isolates collected over about<br />
30 years, we examined M. pneumoniae M129 and<br />
FH for the following features: genome organization,<br />
sequence similarities of selected genes, protein patterns<br />
of two-dimensional gels and transcription profiles.<br />
Comparing the complete nucleotide sequence of the<br />
genome of M.p. M129 with about 150.000 bp of the<br />
genome of M. pneumoniae FH showed that the isolates<br />
are very conserved and so far the main differences<br />
are located in the P1 operon<br />
The P1 operon consists of the following three genes<br />
named ORF4, ORF5 (=P1 gene) and ORF6. The gene<br />
product of ORF6 are the two proteins P40 and P90,<br />
which are involved in the correct localization of P1 in<br />
the membrane. While the ORF4 gene is highly conserved<br />
in both prototypes, the genes P1 and ORF6 are<br />
different. Experiments are in progress to extend the<br />
comparative analyses to other isolates from patients to<br />
examine whether the P1 operon is indeed the decisive<br />
difference between subtypes.<br />
78<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(Graduiertenkolleg “Kontrolle <strong>der</strong> Genexpression<br />
in pathogenen Mikroorganismen”, Projekt-<br />
’Sachbeihilfen’) from the BioRegion Rhein Neckar<br />
(BMBF - Fa. Genzyme Virotech and BMBF - Fa.<br />
EML) and by the Fonds <strong>der</strong> Chemischen Industrie.<br />
PUBLICATIONS<br />
Pyrowolakis, G., Hofmann, D. and Herrmann, R.<br />
(1998). The subunit b of the F 0 F 1 type ATPase of the<br />
bacterium Mycoplasma pneumoniae is a lipoprotein.<br />
J. Biol. Chem 273, 24792-24796.<br />
Herrmann, R. and Reiner, B. (1998). Mycoplasma<br />
pneumoniae – Mycoplasma genitalium, a comparison<br />
of two closely related bacterial species. Curr. Opinion<br />
in Microbiol. 1, 572-579.<br />
Herrmann, R., Göhlmann, H.W.H., Regula, J.T.,<br />
Weiner III, J., Pirkl, E., Ueberle, B. and Frank, R.<br />
(1999). Mycoplasmas, the Smallest Known Bacteria.<br />
In Microbial Evolution and Infection. (U. B. Göbel,<br />
B.R. Ruf, eds.), Einhorn-Presse Verlag, 71-79.<br />
Fisseha, M., Göhlmann, H.W.H., Herrmann, R. and<br />
Krause, D.C. (1999) Identification and Complementation<br />
of Frameshift mutations associated with loss of<br />
cytadherence in Mycoplasma pneumoniae. J. Bacteriol.<br />
181, 4404-4410.<br />
Göhlmann, H.W.H, Weiner, J., Schön, A. and Herr–<br />
mann, R. (<strong>2000</strong>). Identification of a small RNA<br />
within the pdh gene cluster of Mycoplasma pneumoniae<br />
and Mycoplasma genitalium. J. Bacteriol.,<br />
182, 3281-3284.<br />
Dandekar, T., Huynen, M., Regula, J.T., Zimmermann,<br />
C.U., Ueberle, B., Andrade, M., Doerks, T., Sanchez,<br />
L., Snel, B., Suyama, M., Yuan, Y.P., Herrmann, R.,<br />
and Bork, P. (<strong>2000</strong>). Re-annotating the Mycoplasma<br />
pneumoniae genome sequence: Adding value, function<br />
and reading frames. Nucleic Acids Res. (in press).<br />
Regula, J.T., Ueberle, B., Boguth, G., Görg, A.,<br />
Schnölzer, M., Herrmann, R., and Frank, R. (<strong>2000</strong>).<br />
Towards a two-dimensional proteome map of Mycoplasma<br />
pneumoniae. Electrophoresis (in press).<br />
Weiner, J., Herrmann, R., and Browning, G.F. (<strong>2000</strong>).<br />
Transcription in Mycoplasma pneumoniae. Nucleic<br />
Acids Res. (in press).<br />
THESES<br />
Dissertations<br />
Göhlmann, Hinrich (1999). Transcriptionsanalyse<br />
einer gesamten Zelle am Beispiel von Mycoplasma<br />
pneumoniae.<br />
Regula, Jörg (1999). Auf dem Weg zum Proteom von<br />
Mycoplasma pneumoniae.<br />
STRUCTURE OF THE GROUP<br />
E-mail: r.herrmann@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Herrmann, Richard, Prof. Dr.<br />
Research associates Reiser, Walter, Dr.*<br />
Hofmann, Diana, Dipl. Biol.*<br />
Regula, Jörg, Dipl. Chem.<br />
PhD. students Göhlmann, Hinrich, Dipl. Biol.*<br />
Weiner, January, Dipl. Biol.<br />
Catrein, Ina, Dipl. Biol.*<br />
Ueberle, Barbara, Dipl. Biol.<br />
Diploma students C.U. Zimmermann*<br />
Stefan Schulz*<br />
Atcha Boonmee*<br />
Techn. assistants Pirkl, Elsbeth<br />
Götzmann, Martina*<br />
Schaller, Werner*<br />
*part of the time reported<br />
79
Ralf-Peter Jansen<br />
Asymmetric Cell Division and RNA<br />
Transport in Yeast<br />
During embryogenesis, cell type diversity is generated<br />
by different mechanisms, one of which is asymmetric<br />
cell division. Such divisions produce progeny differing<br />
in morphology, size, or gene expression pattern.<br />
However, asymmetric divisions are not only found in<br />
multicellular organisms. A typical unicellular eukaryote<br />
that divides asymmetrically is the yeast Saccharomyces<br />
cerevisiae. A „mother cell“ generates a<br />
„daughter cell“ by polarized growth, a process known<br />
as „budding“. After separation, the two cells show<br />
a differential gene expression pattern. Whereas the<br />
mother cell can express the HO gene and eventually<br />
un<strong>der</strong>goes a process called „mating type switching“,<br />
the daughter cell is unable to do so.<br />
HO expression depends on multiple positively and<br />
negatively acting transcription factors but the differential<br />
expression pattern of HO is regulated by the<br />
asymmetric distribution of a transcriptional repressor<br />
known as Ash1p. Ash1p can be detected in post-anaphase<br />
cells predominantly in the nucleus of daughter<br />
cells where it persists to the end of the daughter cell‘s<br />
following G1 phase. In the daughter cell it is both sufficient<br />
and essential to shut off HO expression in G1.<br />
The asymmetric distribution of Ash1p is the result of<br />
an asymmetric localization of its mRNA during anaphase.<br />
ASH1 mRNA is transported from the nuclei of<br />
both mother and daughter cell to the cell cortex of<br />
the daughter. The localization depends on a functional<br />
microfilament system and three cis-acting localization<br />
signals. These signals appear to be independent of each<br />
other since each signal is sufficient to target a reporter<br />
RNA to the daughter cell. In addition to the cis signals,<br />
5 so called She proteins are essential for ASH1 mRNA<br />
localization. Among these proteins is a type V myosin<br />
80<br />
(She1p/Myo4p) and She5p/Bni1p, a member of the<br />
FH („formin homology“) protein family required for<br />
actin cytoskeleton organization in different organisms.<br />
Homologs of another She protein, She4p have recently<br />
been predicted to be involved in myosin assembly or<br />
function. The last two She proteins, She2p and She3p<br />
do not show any sufficient homology to other proteins.<br />
Myo4p, a motor protein essential for mRNA<br />
transport<br />
D. Djandji, A. Frank, C. Kruse, S. Münchow, and C.<br />
Sauter<br />
To date, Myo4p is the only motor protein with an essential<br />
role in mRNA localization. In or<strong>der</strong> to un<strong>der</strong>stand<br />
its role we initially wanted to test if the myosin<br />
is directly involved in ASH1 mRNA transport.<br />
Using a combination of in-situ hybridization and indirect<br />
immunofluorescence, we could show a colocalization<br />
of the myosin and ASH1 mRNA not only at the<br />
final targeting site (the tip of the daughter cell) but<br />
also on filamentous structures that are most likely microfilaments<br />
running from the mother to the daughter<br />
cell (Fig. 1). In addition, ASH1 mRNA was shown to<br />
specifically coprecipitate with an epitope-tagged version<br />
of Myo4p. Both sets of data strongly suggest that<br />
ASH1 mRNA is in fact a cargo of the Myo4p myosin.<br />
What other proteins are required for the association of<br />
ASH1 mRNA with Myo4p? One candidate was She3p<br />
since it‘s intracellular localization was very similar<br />
to that of Myo4p. We were able to demonstrate that<br />
She3p is essential for Myo4p-ASH1 association. Furthermore,<br />
She3p coprecipitated both with Myo4p and<br />
Figure 1<br />
ASH1 mRNA. In addition, She3p‘s aminoterminus<br />
that is predicted to form a coiled coil can bind to<br />
the carboxyterminal tail of Myo4p in a two hybrid assay<br />
whereas She3p‘s carboxyterminus interacts with<br />
She2p. She3p and She2p might be adapters between<br />
the mRNA and the motor protein.<br />
The role of Myo4p‘s tail domain was further investigated<br />
since the tail domains of other myosins are<br />
involved in cargo binding. Expression of the tail in an<br />
otherwise wildtype cell had the effect of a dominant<br />
negative mutant. It abolished the localization of ASH1<br />
mRNA, She3p and endogenous Myo4p as well as the<br />
association of endogenous Myo4p with She3p, most<br />
likely by sequestering essential factors. The effects are<br />
specific since the expression of another type V myosin<br />
tail did not interfere with mRNA localization.<br />
The myosin tail domain should be useful to biochemically<br />
identify and copurify components of the putative<br />
mRNA transport complex, especially the RNAbinding<br />
factor(s).<br />
Ash1p as a regulator of pseudohyphal differentiation<br />
K. Kahlina and S. Münchow<br />
Besides it‘s role in regulating HO expression and<br />
mating type switching, Ash1p is required for another<br />
differentiation process in Saccharomyces cerevisiae,<br />
pseudohyphal growth. During this differentiation,<br />
yeast cells elongate and grow unipolar to form chains<br />
of connected cells that allow a yeast colony to penetrate<br />
it‘s medium. Several signalling pathways converge<br />
during induction of pseudohyphal growth and<br />
at least four transcription factors (including Ash1p)<br />
are required for this differentiation process. However,<br />
nothing is known about the signalling pathway that<br />
regulates Ash1p or about the target genes that are regulated<br />
by Ash1p.<br />
We have recently shown that mislocalization of ASH1<br />
mRNA and Ash1 protein in a myo4 mutant dramatically<br />
increases pseudohyphal growth suggesting that<br />
RNA targeting plays a crucial role in proper pseudohyphal<br />
differentiation. In or<strong>der</strong> to identify ASH1-dependent<br />
genes, two independent screens based on dif-<br />
81
ferential display methods and gene arrays have been<br />
initiated and putative candidates are currently analyzed.<br />
Pseudohyphal and hyphal growth plays an important<br />
role during pathogenesis of the yeast Candida albicans.<br />
In the course of the C. albicans sequencing project,<br />
a DNA fragment of the putative C. albicans ASH1<br />
gene has been identified. Using this fragment, we have<br />
cloned the full length C. albicans ASH1 gene and are<br />
currently characterizing its function.<br />
Novel localized mRNAs in Saccharomyces<br />
cerevisiae<br />
F. Böhl and D. Ferring<br />
In a number of cell types, several mRNAs are sorted<br />
differentially in a single cell. In or<strong>der</strong> to identify novel<br />
localized RNAs in yeast, we devised two different<br />
screens based on in-situ hybridization. The first one<br />
was based on the rationale that asymmetrically sorted<br />
mRNAs should be expressed in the same cell cycle<br />
window as ASH1, the M-/G1-phase boundary. Therefore,<br />
we fused the 3‘ parts of 40 mRNAs that are expressed<br />
at this boundary to a reporter mRNA (GFP,<br />
green fluorescent protein) and expressed these hybrids<br />
from a regulatable promoter. In the case of ASH1 such<br />
an RNA hybrid is clearly localized to the daughter cell.<br />
However, none of the other mRNA hybrids showed a<br />
localized hybridization signal, indicating that ASH1 is<br />
the only M-/G1-phase mRNA that is asymmetrically<br />
distributed.<br />
In a second approach, we tempted to screen the complete<br />
yeast genome for localized mRNAs. A plasmidbased<br />
DNA library was generated that allowed random<br />
fusions of genomic DNA fragments to the GFP coding<br />
sequence. The resulting hybrid genes can be expressed<br />
from a regulatable GAL1 promoter. Localized RNAs<br />
82<br />
can be detected by in-situ hybridization against the<br />
GFP part of the resulting hybrid mRNAs. The constructed<br />
library covers approximately 95% of the complete<br />
yeast genome. After transformation into yeast,<br />
pools of 25 clones are currently analyzed for the presence<br />
of novel localized mRNAs. Such a screen should<br />
be useful to assess the complete number of localized<br />
mRNAs in a polarized cell.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (Graduiertenkolleg “<strong>Molekulare</strong> Zellbiologie”,<br />
Projekt-’Sachbeihilfen’).<br />
PUBLICATIONS<br />
Münchow, S., Sauter, C., and Jansen, R.-P. (1999). Association<br />
of the class V myosin Myo4p with a localised<br />
messenger RNA in budding yeast depends on She proteins.<br />
J. Cell Sci. 112, 1511-1518.<br />
Jansen, R.-P. (1999). RNA-cytoskeletal associations.<br />
FASEB J. 13, 455-466.<br />
Hurt, E., Segref, A., Sträßer, K., Bailer, S., Schlaich,<br />
N., Presutti, C., Tollervey, D., and Jansen, R.-P. (<strong>2000</strong>).<br />
Mex67p mediates nuclear export of a variety of Pol II<br />
transcripts. J.Biol.Chem. 275, 8361-8368.<br />
THESES<br />
Diploma<br />
Münchow, S. (1998). Isolierung und Charakterisierung<br />
des Myo4-Protein/ASH1 mRNA Komplexes aus Sac-<br />
charomyces cerevisiae.<br />
Sauter, C. (1998). Detektion lokalisierter mRNAs in<br />
Saccharomyces cerevisiae mittels Fluoreszenz-in situ-<br />
Hybridisierung.<br />
Theurer, J. (1998). Versuch <strong>der</strong> Identifikation neuer<br />
Bestandteile <strong>der</strong> ASH1 mRNA Lokalisationsmaschi–<br />
nerie von Saccharomyces cerevisiae durch Überexpression<br />
einer cDNA-Bibliothek.<br />
Djandji, D. (1999). Die Assoziation des She3 Proteins<br />
mit ASH1 mRNA in Saccharomyces cerevisiae.<br />
STRUCTURE OF THE GROUP<br />
e-mail: r.jansen@mail.zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Jansen, Ralf-Peter, Dr.<br />
Ph.D. students Böhl, Florian, Dipl. Biol. *<br />
Jaedicke, Andreas, Dipl. Biol. *<br />
Kruse, Claudia, Dipl. Biol. *<br />
Münchow, Sonja, Dipl. Biol.<br />
Diploma students Djandji, Dominic *<br />
Sauter, Claus *<br />
Theurer, Jörg *<br />
Kahlina, Kornelia *<br />
Techn. assistants Ferring, Dunja *<br />
Frank, Andrea *<br />
* part of the time reported<br />
83
Stefan Jentsch<br />
Ubiquitin-Dependent Proteolysis<br />
The central importance of selective proteolytic systems<br />
in regulating cellular events has been recognized<br />
recently. Progression through the eukaryotic cell<br />
cycle, for example, is substantially regulated through<br />
a timed and coordinated degradation of cyclins and<br />
inhibitors of cyclin-dependent protein kinases. Similarly,<br />
the shift from one transcriptional or developmental<br />
program to another is often achieved through<br />
a timed destruction of regulatory proteins. Proteolysis<br />
is irreversible and therefore proteolytic enzymes are<br />
usually employed for controlling unidirectional cellular<br />
pathways. Selective degradation in eukaryotes is<br />
primarily mediated by the ubiquitin/proteasome pathway.<br />
Substrates of this pathway are covalently modified<br />
by conjugation to ubiquitin, a small and highly<br />
conserved protein. In most cases several ubiquitin<br />
molecules are added to the substrate as a multiubi–<br />
quitin chain in which ubiquitin molecules are arranged<br />
like beads on a string. Multiubiquitinated proteins are<br />
targeted for degradation by a large protease complex,<br />
known as the 26S proteasome. Our research focuses<br />
primarily on functional aspects of this pathway. We<br />
identify from yeast and mammalian cells the enzymatic<br />
components of this system, clone the corresponding<br />
genes, and study their functions in vitro and<br />
in vivo.<br />
A novel ubiquitination factor, E4, involved in<br />
multiubiquitin chain assembly<br />
M. Kögl, T. Hoppe, S. Schlenker, H. D. Ulrich, T. U.<br />
Mayer<br />
Proteins modified by multiubiquitin chains are the<br />
preferred substrates of the proteasome. Previous work<br />
has suggested that E1, E2, and E3 enzymes are both<br />
84<br />
required and sufficient for the formation of multi–<br />
ubiquitinated substrates. Recently, however, we could<br />
show that efficient multiubiquitination needed for proteasomal<br />
targeting of a model substrate (ubiquitin<br />
fusions) requires an additional conjugation factor,<br />
which we termed E4. This protein, previously known<br />
as UFD2 in yeast, binds to the ubiquitin moieties of<br />
preformed conjugates and catalyzes ubiquitin chain<br />
assembly in conjunction with E1, E2, and E3. Intriguingly,<br />
E4 defines a new protein family, which includes<br />
two human members and the regulatory protein NOSA<br />
from Dictyostelium required for fruiting body development.<br />
In yeast E4 activity is linked to cell survival<br />
un<strong>der</strong> stress conditions, indicating that eukaryotes utilize<br />
E4-dependent proteolysis pathways for multiple<br />
cellular functions. Interestingly, UFD2 (E4) interacts<br />
with CDC48, a member of the large family of AAAtype<br />
ATPases. CDC48 is not involved in the ubiquitinconjugation<br />
reaction but is needed for proteolysis of<br />
some substrates of the ubiquitin/proteasome pathway.<br />
Ubiquitin-like proteins from mammals and<br />
yeast<br />
D. Liakopoulos, G. Doenges<br />
Ubiquitin is one of the most highly conserved eukaryotic<br />
proteins known to date. Human ubiquitin differs<br />
from its yeast homolog by only three amino acid residues<br />
(out of 76 residues) and the proteins are functionally<br />
equivalent. In addition to ubiquitin, two classes<br />
of ubiquitin-related proteins have been identified. Proteins<br />
of the first class carry ubiquitin-like domains<br />
linked to unrelated sequences and are not conjugated<br />
to other cellular proteins. One example is the mammalian<br />
ubiquitin-related protein BAG-1 for which we<br />
showed that it functions as a regulatory cofactor of<br />
the Hsc70 chaperone. Proteins of the second class of<br />
ubiquitin-like proteins are distinguished by their property<br />
to become posttranslationally attached to other<br />
cellular proteins analogously to ubiquitin. Known<br />
members are SUMO-1 from higher eukaryotes and<br />
its yeast ortholog SMT3. Recently, we have identified<br />
a novel ubiquitin-like protein from yeast, which<br />
we termed RUB1. RUB1 displays 53% amino acid<br />
sequence identity to ubiquitin. We found that RUB1<br />
conjugation to other cellular proteins requires at least<br />
three proteins in vivo. ULA1 and UBA3 are related<br />
to the amino- and carboxyl-terminal domains of the<br />
E1 ubiquitin-activating enzyme, respectively, and together<br />
fulfill E1-like functions for RUB1 activation.<br />
RUB1 conjugation also requires UBC12, a protein<br />
related to E2 ubiquitin-conjugating enzymes, which<br />
functions analogously to E2 enzymes in RUB1-protein<br />
conjugate formation. Conjugation of RUB1 is not<br />
essential for normal cell growth and appears to be<br />
selective for a small set of substrates. Remarkably, we<br />
found that CDC53/cullin, a common subunit of the<br />
multifunctional SCF ubiquitin ligase, is a major substrate<br />
for RUB1 conjugation. This suggests that the<br />
RUB1-conjugation pathway is functionally affiliated<br />
with the ubiquitin/proteasome system and may play a<br />
regulatory role.<br />
Conjugation of the ubiquitin-like protein RUB1/<br />
NEDD8 to cullin-2 is linked to von Hippel-Lindau<br />
(VHL) tumor suppressor function<br />
D. Liakopoulos, G. Doenges; in collaboration with T.<br />
Büsgen, A. Brychzy, and A. Pause (Max Planck Institute<br />
for Biochemistry, Martinsried)<br />
Yeast RUB1 has homologs in higher eukaryotic cells.<br />
The human RUB1 homolog, NEDD8, when expressed<br />
in a yeast rub1 mutant, can complement its RUB1<br />
deficiency. Recently we found that both hCUL-1<br />
and hCUL-2 (homologs of yeast CDC53, see above)<br />
are modified by the conserved ubiquitin-like protein<br />
RUB1/NEDD8. Whereas hCUL-1 is part of a human<br />
SCF complex, hCUL-2 assembles with elongin B/C<br />
and the von Hippel-Lindau tumor suppressor protein<br />
pVHL, forming a protein complex, CBC VHL , that<br />
resembles SCF ubiquitin ligases. We could show that<br />
NEDD8-hCUL-2 conjugates are part of CBC VHL complexes<br />
in vivo. Remarkably, the formation of these<br />
conjugates is stimulated by the pVHL tumor suppressor.<br />
A tumorigenic pVHL variant, however, is<br />
essentially deficient in this activity. Thus, ligation of<br />
NEDD8 to hCUL-2 is linked to pVHL activity and<br />
may be important for pVHL tumor suppressor function.<br />
Moreover, our data indicate that modification of<br />
cullins by NEDD8 requires the existence of a preassembled<br />
ubiquitin ligase complex.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from Deutsche Forschungsgemeinschaft<br />
(project grants, SFB 352, Schwerpunktprogramm<br />
Ubiquitin/Proteasomsystem, Leibniz Program,<br />
Graduiertenkolleg), European TMR Ubiquitin<br />
Research Network, American Cancer Society, and<br />
Fonds <strong>der</strong> Chemischen Industrie.<br />
PUBLICATIONS<br />
Finley et al., (1998). Unified nomenclature for subunits<br />
of the Saccharomyces cerevisiae proteasome regulatory<br />
particle. Trends Biochem. Sci. 23, 244-245.<br />
Hauser, H.-P., Bardroff, M., Pyrowolakis, G., and<br />
85
Jentsch, S. (1998). A giant ubiquitin-conjugating<br />
enzyme related to IAP apoptosis inhibitors. J. Cell<br />
Biol. 141, 1415-1422.<br />
Jentsch, S. and Ulrich, H.D. (1998). Ubiquitous déjà<br />
vu. Nature 393, 321-323.<br />
Liakopoulos, D., Doenges, G., Matuschewski, K., and<br />
Jentsch, S. (1998). A novel protein modification pathway<br />
related to the ubiquitin system. EMBO J. 17,<br />
2208-2214.<br />
Mayer, T.U., Braun, T., and Jentsch, S. (1998). Role of<br />
the proteasome in membrane extraction of a short-lived<br />
ER-transmembrane protein. EMBO J. 17, 3251-3257.<br />
Scheffner, M., Smith, S., and Jentsch, S. (1998).<br />
The ubiquitin-conjugation system. In: Ubiquitin. J.M.<br />
Peters, J.R. Harris , and D. Finley, eds., Plenum Press,<br />
New York, pp 65-98.<br />
Schwarz, S.E., Matuschewski, K., Liakopoulos, D.,<br />
Scheffner, M., and Jentsch, S. (1998). The ubiquitinlike<br />
proteins SMT3 and SUMO-1 are conjugated by<br />
the UBC9 E2 enzyme. Proc. Natl. Acad. Sci. USA 95,<br />
560-564.<br />
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H.D.,<br />
Mayer, T.U., and Jentsch, S. (1999). A novel ubiquitination<br />
factor, E4, is involved in multiubiquitin chain<br />
assembly. Cell 96, 635-644.<br />
Liakopoulos, D., Büsgen, T., Brychzy, A., Jentsch, S.,<br />
and Pause, A. (1999). Conjugation of the ubiquitinlike<br />
protein NEDD8 to cullin-2 is linked to von Hippel-Lindau<br />
(VHL) tumor suppressor function. Proc.<br />
Natl. Acad. Sci. USA 96, 5510-5515.<br />
86<br />
Honors/Awards<br />
Elected Member of the German Academy of Science,<br />
Leopoldina, 1998.<br />
Member of the Advisory Editorial Board of EMBO<br />
Journal, 1998.<br />
Member of the Advisory Editorial Board of EMBO<br />
Reports, 1999.<br />
THESES<br />
Dissertations<br />
Braun, T. (1998). Analyse des Abbaus von Membranproteinen<br />
am Endoplasmatischen Retikulum.<br />
Liakopoulos, D. (1998). Identifizierung eines neuen<br />
Protein-Modifikations-Systems mit Verwandtschaft<br />
zum Ubiquitin-System.<br />
Matuschewski, K. (1998). Genetische Charakterisie–<br />
rung <strong>der</strong> Ubiquitin-Protein-Ligase RSP5.<br />
Mayer, T. (1998). Rolle des Proteasoms bei <strong>der</strong><br />
Extraktion eines kurzlebigen Proteins aus <strong>der</strong> ER-<br />
Membran.<br />
STRUCTURE OF THE GROUP<br />
Group lea<strong>der</strong>: Jentsch, Stefan, Prof. Dr.<br />
Postdoctoral<br />
fellows: Ulrich, Helle, Ph.D.<br />
Kögl, Manfred, Dr.<br />
Ph.D. students:<br />
Bardroff, Michael, Dipl. Biochem.<br />
Braun, Thorsten, Dipl. Chem.<br />
Doenges, Georg, Dipl. Biol.<br />
Hoege, Carsten, Dipl. Biol., M. Phil.<br />
Hoppe, Thorsten, Dipl. Biol.<br />
Liakopoulos, Dimitris, Dipl. Chem.<br />
Matuschewski, Kai, Dipl. Biochem.<br />
Mayer, Thomas, Dipl. Biol.<br />
Pyrowolakis, George, Dipl. Biol.<br />
Thoms, Sven, Dipl. Chem.<br />
Schlenker, Stephan, Dipl. Biochem.<br />
Technical<br />
assistants: Hubbe, Petra<br />
Jepsen, Kathrin<br />
Löser, Eva<br />
87
Project Group Jörg Höhfeld<br />
Function and Regulation of the Mammalian<br />
Chaperone Hsc70<br />
We are focusing on the characterization of the Hsc70<br />
chaperone system in the mammalian cytosol and<br />
nucleus. Hsc70 is not only involved in the folding of<br />
newly-synthesized polypeptides but also in the sorting<br />
of proteins to different cellular compartments and<br />
appears to present proteins to the degradative system<br />
of the cell. Thus, elucidating the regulation of Hsc70<br />
will provide insight into central events during protein<br />
biogenesis. In recent years we have been able to<br />
identify and characterize several chaperone cofactors<br />
that modulate the chaperone activity of Hsc70. Thus,<br />
essential tools have become available to decipher the<br />
molecular mechanisms that determine the functional<br />
specificity of Hsc70 and that enable Hsc70 to efficiently<br />
cooperate with other chaperone systems and<br />
with the proteolytic machinery.<br />
Structural requirements for the interaction of<br />
Hsc70 with distinct cofactors<br />
J. Demand<br />
The modulation of the chaperone activity of the heat<br />
shock cognate Hsc70 protein in mammalian cells<br />
involves a cooperation with chaperone cofactors such<br />
as Hsp40, BAG-1, the Hsc70-interacting protein Hip,<br />
and the Hsc70/Hsp90-organizing protein Hop. By<br />
employing the yeast two-hybrid system and in vitro<br />
interaction assays we were able to determine structural<br />
requirements for Hsc70‘s cooperation with different<br />
cofactors. The carboxy-terminal domain of Hsc70,<br />
previously shown to form a lid over the peptide bind-<br />
88<br />
ing pocket of the chaperone protein, mediates interaction<br />
of Hsc70 with Hsp40 and Hop. Remarkably, the<br />
two cofactors bind to the carboxy terminus of Hsc70<br />
in a non-competitive manner, revealing the existence<br />
of distinct binding sites for Hsp40 and Hop within this<br />
domain. In contrast, Hip exclusively interacts with<br />
the amino-terminal ATPase domain of Hsc70. Hence,<br />
Hsc70 possesses separate non-overlapping binding<br />
sites for Hsp40, Hip, and Hop. This appears to enable<br />
the chaperone protein to cooperate simultaneously<br />
with multiple cofactors. On the other hand, BAG-1<br />
and Hip have recently be shown to compete in bind-<br />
Figure 1. Cofactor-binding sites of Hsc70<br />
ing to the ATPase domain. Our data thus establish the<br />
existence of a network of cooperating and competing<br />
cofactors regulating the chaperone activity of Hsc70<br />
in the mammalian cell.<br />
Cofactor-induced modulation of the functional<br />
specificity of the molecular chaperone Hsc70<br />
J. Lü<strong>der</strong>s<br />
Molecular chaperones differ in their ability to stabilize<br />
nonnative polypeptides and to mediate protein<br />
folding, defining ‚holding‘ and ‚folding‘ systems. We<br />
analyzed how the chaperone activity of Hsc70 is modulated<br />
by different cofactors after in vitro reconstitu-<br />
tion of the chaperone system. It was observed that<br />
Hsc70 in conjunction with the cofactor Hsp40 stabilizes<br />
heat-denatured firefly luciferase, acting as a<br />
‚holding‘ system. The stabilizing activity turns into<br />
a folding activity in the additional presence of the<br />
Hsc70-interacting protein Hip. In contrast, the cofactor<br />
BAG-1 abrogates the ‚holding‘ function of the<br />
Hsc70/Hsp40 system and blocks the action of Hip on<br />
Hsc70. The competing cofactors Hip and BAG-1 thus<br />
directly determine the fate of a nonnative polypeptide<br />
stabilized by Hsc70 and Hsp40. The findings reveal<br />
on a molecular level how functional specificity of<br />
Hsc70 can be induced through the action of chaperone<br />
cofactors.<br />
Figure 2. Cofactors determine the function of Hsc70<br />
Defining a pathway that links molecular chaperones<br />
to the proteolytic machinery<br />
J. Höhfeld<br />
It is intriguing that Hsc70 participates in protein folding<br />
but can also present non-native proteins to the<br />
ubiquitin/proteasome system, which mediates protein<br />
breakdown. Regulatory mechanisms might exist to<br />
modulate the action of Hsc70 at the interface between<br />
protein folding and protein degradation.<br />
Remarkably, the Hsc70 cofactor BAG-1 possesses a<br />
ubiquitin-like domain at its amino-terminus, suggesting<br />
a link to the proteolytic system. To verify this<br />
notion we analyzed whether BAG-1 stably associates<br />
with the proteasome. An association of BAG-1 with<br />
the proteolytic complex was indeed observed in<br />
human HeLa cells. Binding of the chaperone cofactor<br />
to the proteolytic complex is regulated by ATP-hydrolysis<br />
and is not mediated by Hsc70. Intriguingly, targeting<br />
of BAG-1 to the proteasome promotes an association<br />
of Hsc70 with the proteaseome in vitro and<br />
in vivo. BAG-1 apparently acts as a coupling factor<br />
between the chaperone system and the proteolytic<br />
complex. BAG-1 may thus fulfill a key regulatory<br />
function at the interface of protein folding and protein<br />
degradation.<br />
Figure 3. BAG-1 acts as a coupling factor between<br />
Hsc70 and the proteasome<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (Sachbeihilfen).<br />
89
PUBLICATIONS<br />
Heyrovska, N., Frydman, J., Höhfeld, J., and Hartl,<br />
F.-U. (1998). Directionality of polypeptide transfer<br />
in the mitochondrial pathway of chaperone-mediated<br />
protein folding. Biol. Chem. 379, 301-309.<br />
Höhfeld, J. (1998). Regulation of the heat shock cognate<br />
Hsc70 in the mammalian cell: the characterization<br />
of the anti-apoptotic protein BAG-1 provides<br />
novel insights. Biol. Chem. 379, 269-274.<br />
Demand, J., Lü<strong>der</strong>s, J., and Höhfeld, J. (1998). The<br />
carboxy-terminal domain of Hsc70 provides binding<br />
sites for a distinct set of chaperone cofactors. Mol.<br />
Cell. Biol. 18, 2023-2028.<br />
Lü<strong>der</strong>s, J., Demand, J., Schönfel<strong>der</strong>, S., Frien, M.,<br />
Zimmermann, R., and Höhfeld, J. (1998). Cofactorinduced<br />
modulation of the functional specificity of<br />
the molecular chaperone Hsc70. Biol. Chem. 379,<br />
1217-1226.<br />
Lü<strong>der</strong>s, J., Demand, J., and Höhfeld, J. (1999). The<br />
ubiquitin-related BAG-1 provides a link between the<br />
molecular chaperones Hsc70/Hsp70 and the proteasome.<br />
J. Biol. Chem, 275, 4613-4617.<br />
Habilitation<br />
Höhfeld, J. (1998). Regulation of molecular chaperones<br />
in the eukaryotic cell.<br />
STRUCTURE OF THE GROUP<br />
Group lea<strong>der</strong> Höhfeld, Jörg, PD Dr.<br />
Ph.D. students Demand, Jens, Dipl.Chem.<br />
Lü<strong>der</strong>s, Jens, Dipl.Biol.<br />
90<br />
Klaus-Armin Nave<br />
Myelin Genetics and Developmental<br />
Neurobiology<br />
The assembly of myelin in the nervous system of<br />
higher vertebrates is the highly specialized function of<br />
oligodendrocytes and Schwann cells. Myelin-forming<br />
glial cells enwrap axons with multiple layers of<br />
membranes and provide the electrical insulation that<br />
is necessary for a rapid impulse propagation. We<br />
are interested in the principles of these neuron-glia<br />
interactions and are studying genes that are required<br />
for normal myelin assembly and maintenance. Transgenic<br />
and natural mouse mutants are useful tools<br />
which also serve as models for corresponding myelin<br />
diseases in human. One gene of interest encodes<br />
PMP22, a myelin membrane protein of Schwann<br />
cells, and is frequently duplicated in human patients<br />
with Charcot-Marie-Tooth disease. Mutations of the<br />
proteolipid protein (PLP) gene un<strong>der</strong>lie Pelizaeus-<br />
Merzbacher disease, a lethal white matter disease.<br />
Proteolipids are not restricted to glial cells: PLPhomologous<br />
genes (M6A, M6B) are expressed in<br />
nearly all CNS neurons but their function is still<br />
unknown. Mouse mutants of these genes and the overexpression<br />
of proteolipids in heterologous cells shed<br />
some light onto their cellular function.<br />
In an attempt to identify regulatory genes that control<br />
the terminal differentiation of neuronal and glial cells,<br />
we have cloned several basic helix-loop-helix (bHLH)<br />
transcription factors from the postnatal rodent brain.<br />
One bHLH factor that we termed NEX defines a<br />
new subfamily of neuronal bHLH genes expressed<br />
in the mammalian CNS. This familiy of Drosophila<br />
atonal-related genes also includes BETA2/NeuroD<br />
and NDRF. The targeted inactivation of NeuroD and<br />
NEX in transgenic mice reveals a critical function of<br />
these genes in the differentiation program of hippocampal<br />
neurons.<br />
I. Myelin protein PMP22 in a rat model of<br />
Charcot-Marie-Tooth disease<br />
S. Niemann, M. Sereda<br />
Tetraspan myelin proteins play an important role in<br />
CNS and PNS myelination. To address the role of<br />
PMP22 in development and in disease, we have generated<br />
a transgenic rat model of the most frequent<br />
human neuropathy, Charcot-Marie-Tooth disease type<br />
1A. CMT1A is associated with a partial duplication<br />
of chromosome 17 and we have proven experimentally<br />
that the un<strong>der</strong>lying cause of CMT1A is overexpression<br />
of the PMP22 gene. Transgenic rats have<br />
typical clinical features of the human disease, including<br />
muscle weakness, reduced nerve conduction<br />
velocities, and marked Schwann cell hypertrophy. In<br />
the phenotypical analysis of the CMT rat we can now<br />
adress questions which could previously not be studied<br />
in the corresponding human disease. Of specific<br />
interest is the analysis of homozygous PMP22-transgenic<br />
rats which completely fail to elaborate a myelin<br />
sheath. The severalfold increased PMP22 gene dosage<br />
results in a remarkable „uncoupling“ of normal molecular<br />
parameters of Schwann cell differentiation which<br />
coincide with a premature myelination arrest. The<br />
abnormal „gain of function“ effect places the role of<br />
PMP22 upstream in the myelin assembly process.<br />
II. Proteolipid protein in myelin assembly<br />
and dysmyelinating disease<br />
M. Jung, E. Krämer, M. Klugmann<br />
PLP is a highly conserved, myelin-specific protein<br />
with four transmembrane domains. We have previously<br />
identified mutations in the mouse PLP gene<br />
which differ in phenotype depending on the nature of<br />
the specific molecular defect. There is good evidence<br />
that mutations of the PLP gene are lethal because the<br />
91
expression of misfolded PLP is „toxic“ to myelinforming<br />
oligodendrocytes. Jimpy mice can not be<br />
„rescued“ by transgenic complementation with a wildtype<br />
PLP transgene. Moreover, mice which completely<br />
lack expression of PLP or any truncated<br />
polypeptide (a null allele created by gene targeting)<br />
develop normal motor functions and show no signs<br />
of abnormal glial cell death. In fact, CNS myelin is<br />
assembled as a multilamellar and compacted structure<br />
in the absence of PLP. Although the overall stability<br />
of such PLP-deficient myelin appears reduced, these<br />
observations question the view that the major mem-<br />
Figure 1. Mutations of the gene for myelin proteolipid<br />
protein (PLP) cause Pelizaeus-Merzbacher Disease and<br />
severe dysmyelination in PLP-transgenic mice. The molecular<br />
pathomechanism, which is associated with abnormal<br />
protein trafficking, is poorly un<strong>der</strong>stood. We are using confocal<br />
microscopy to study the intracellular localisation and<br />
export of myelin proteins into the processes of cultured oligodendrocytes.<br />
The folding-sensitive O10-epitope of PLP<br />
(in green) which is lacking in all mutant PLP isoforms<br />
emerges after the C-terminal epitope (in red) of newly synthezised<br />
PLP (provided by E. Krämer).<br />
92<br />
brane protein of CNS myelin is essential to obtain<br />
myelin membrane adhesion and compaction. Recent<br />
evidence suggest that PLP deficiency has profound<br />
effects on axonal integrity which becomes clinically<br />
manifest in aged mice with signs of „neuroaxonal<br />
dystrophy“. The basis of the un<strong>der</strong>lying axon-glia<br />
interaction is currently un<strong>der</strong> investigation.<br />
We have expressed PLP cDNAs from natural mouse<br />
mutants in cultured non-glial cells. These studies show<br />
that fibroblasts are much less sensitive to the overexpression<br />
effect and mutant isoforms of PLP than<br />
oligodendrocytes in vivo. However, also transfected<br />
fibroblastoid cells recognize subtle alterations in the<br />
PLP primary structure, because these translation products<br />
are retained inside the cell and do not reach the<br />
cell surface. A direct demonstration of protein misfolding<br />
is also possible with a conformation-sensitive<br />
monoclonal antibody (O10) which distinguishes<br />
wildtype PLP from the known mutant isoforms.<br />
III. Neuronal members of the proteolipid protein<br />
family: M6A and M6B<br />
L. Dimou, M. Klugmann, H. Werner<br />
PLP is the prototype of a small family of sequencerelated<br />
tetraspan membrane proteins which includes<br />
M6A and M6B proteins, abundantly expressed on<br />
neuronal processes in the central nervous system.<br />
M6B is also detectable in oligodendrocytes. There are<br />
multiple M6B-mRNAs, encoding at least eight protein<br />
isoforms. Smaller, presumably soluble polypeptides<br />
result from alternatively spliced M6B mRNAs<br />
with a stop codon upstream of the exon for the first<br />
transmembrane domain. In cells of the oligodendroglial<br />
lineage, M6B localizes to the cell surface and transiently<br />
also on cellular processes. By in situ-hybrid-<br />
ization, oligodendrocytes express one of two alternative<br />
C-termini, whereas cortical neurons express<br />
both. Regulated expression of the various intracellular<br />
domains of M6B, termed α, β, γ, ψ, and ω (some of<br />
which are structurally conserved in M6A and PLP),<br />
suggests that proteolipids can interact with cytoplasmic<br />
proteins present in a variety of neural cell types.<br />
The N-terminal domains of M6B affect transport features<br />
and the subcellular distribution of this protein<br />
when expressed in fibroblasts. The analysis of M6A<br />
and M6B mouse mutants is ongoing and suggests a<br />
role in neuronal process outgrowth and CNS myelination,<br />
respectively.<br />
IV. NEX and NeuroD regulate hippocampal<br />
granule cell differentition<br />
A. Bartholomä, S. Göbbels, M. Rossner, M.<br />
Schwab<br />
Parts of our laboratory‘s activity was devoted to the<br />
function of basic helix-loop-helix (bHLH) proteins in<br />
the nervous system. The neuronal transcription factors<br />
NEX (neuronal helix-loop-helix protein-1), NeuroD<br />
(neurogenic differentiation factor), and NDRF comprise<br />
a family of Drosophila atonal-related bHLH<br />
proteins with highly overlapping expression in the<br />
developing forebrain. A role for NeuroD and NEX<br />
in terminal neuronal differentiation is demonstrated<br />
by mutations in mice. For example, in the hippocampus,<br />
presumptive granule cells of the dentate gyrus<br />
are generated but fail to mature, lack normal sodium<br />
currents, and show little dendritic arborization. Longterm<br />
hippocampal slice cultures reveal secondary<br />
alterations, such as abnormal projections from the<br />
entorhinal cortex. Ongoing experiments aim at restricting<br />
the mutations of the NeuroD gene to subsets of<br />
cortical neurons, utilizing homologous recombination<br />
of the loxP/Cre system. This will allow to circumvent<br />
perinatal lethality and study the role of bHLH proteins<br />
in adult neuronal functions.<br />
Figure 2. Cre-mediated recombination of genes that are<br />
flanked by loxP sites allows the generation of cell type-specific<br />
somatic mutations in transgenic mice. We have utilized<br />
the spatio-temporal expression pattern of the NEX gene<br />
to drive expression of Cre specifically into CNS neurons. A<br />
„knock in“ strategy was chosen, to insert Cre into exon 2 of<br />
the endogenous NEX gene. To demonstrate the specificity of<br />
this system, a lacZ reporter function was specifically activated<br />
by Cre recombination in neurons of the hippocampus<br />
and neocortex of transgenic mice. This mouse is used to<br />
circumvent the perinatal lethality of a null mutation in the<br />
neuroD gene (provided by S. Goebbels).<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(SFB 317 “<strong>Molekulare</strong> <strong>Biologie</strong> neuraler Mechanismen<br />
und Interaktionen”, Graduiertenkolleg “<strong>Molekulare</strong><br />
und zelluläre Neurobiologie”) from the BMBF<br />
and from the EU.<br />
93
PUBLICATIONS<br />
An<strong>der</strong>son, T.J., Klugmann, M., Thomson, C.,<br />
Schnei<strong>der</strong>, A., Readhead, C., Nave, K.-A., and<br />
Griffiths, I.R. (1999). Distinct phenotypes associated<br />
with increasing dosage of the PLP gene: implications<br />
for gene duplications in CMT1A. Annals N.Y. Acad.<br />
Sci. 883, 234-236.<br />
An<strong>der</strong>son, T.J., Schnei<strong>der</strong>, A., Barrie, J.A., Klugmann,<br />
M., McCulloch, M.C., Kirkham, D., Kyriakides, E.,<br />
Nave, K.-A. and Griffiths, I.R. (1998). Increased<br />
dosage of the proteolipid protein gene causes late-onset<br />
neurodegeneration. J. Comp. Neurol. 394, 506-519.<br />
Bradl, M., Bauer, J., Inomata, T., Zielasek, J., Nave,<br />
K.-A., Toyka, K., Lassmann, H., and Wekerle, H.<br />
(1999). Transgenic Lewis rats overexpressing the proteolipid<br />
protein gene: myelin degeneration and its<br />
effect on T cell-mediated experimental autoimmune<br />
encephalomyelitis. Acta Neuropathol. 97, 595-606.<br />
Coetzee, T., Suzuki, K., Nave, K.-A., and Popko,<br />
B. (1999). Myelination in the absence of galactolipids<br />
and proteolipid proteins. Mol. Cell. Neurosci. 14,<br />
41-51.<br />
Dimou, L., Klugmann, M., Werner, H., Jung, M.,<br />
Griffiths, I.R., and Nave, K.-A. (1999). Dysmyelination<br />
in mice and the proteolipid protein gene family.<br />
Adv. Exp. Med. Biol. 468, 261-271.<br />
Griffiths, I., Klugmann, M., An<strong>der</strong>son, T. and Nave,<br />
K.-A. (1998). Current concepts of PLP and its role<br />
in the nervous system. Microscop. Res. Techn. 41,<br />
344-358.<br />
94<br />
Griffiths, I., Klugmann, M., An<strong>der</strong>son, T., Yool, D.,<br />
Thomson, C., Schwab, M.H., Schnei<strong>der</strong>, A., Zimmermann,<br />
F., McCulloch, M., Nadon, N., and Nave,<br />
K.-A. (1998). Axonal swellings and degeneration in<br />
mice lacking myelin proteolipid protein. Science 280,<br />
1610-1613.<br />
Klein, L., Klugmann, M., Nave, K.-A., and Kyewski,<br />
B. (1999). Shaping of the autoreactive T-cell repertoire<br />
by a splice variant of self protein expressed in<br />
thymic epithelial cells. Nature Medicine 6, 56-61.<br />
Montague, P., Kirkham, D., McCallion, A.S., Davies,<br />
R.W., Kennedy, P.G., Klugmann, M., Nave, K.-A.,<br />
and Griffiths, I.R. (1999). Reduced levels of a specific<br />
myelin-associated oligodendrocytic basic protein isoform<br />
in shiverer myelin. Dev. Neurosci. 21, 36-42.<br />
Nave, K.-A. (1999) X-linked dysmyelination: mouse<br />
models of Pelizaeus-Merzbacher disease. In Mouse<br />
models in the study of genetic neurological disor<strong>der</strong>s.<br />
B. Popko, ed. (Plenum Press, New York), pp 25-41.<br />
Nave, K.-A. (1999). Zentrale Myelinisierungsstörungen:<br />
Biologische Grundlagen, transgene Modelle und<br />
molekulare Pathologie. In Handbuch <strong>der</strong> <strong>Molekulare</strong>n<br />
Medizin, Band V. D. Ganten and K. Ruckpaul, eds.<br />
(Springer Verlag Berlin), pp 370-394.<br />
Nave, K.-A. and Trapp, B. (<strong>2000</strong>). Myelin-forming<br />
glial cells. Glia 29, 103.<br />
Nave, K.-A. (<strong>2000</strong>). Myelin-specific genes and their<br />
mutations in the mouse. In Glial Cell Development,<br />
2 nd edition. K.R. Jessen and W.D. Richardson, eds.<br />
(Oxford University Press), (in press).<br />
Niemann, S., Sidman, R.L. and Nave, K.-A. (1998).<br />
Evidence against altered forms of MAG in the dysmyelinated<br />
mouse mutant claw paw. Mammalian<br />
Genome 9, 903-904.<br />
Niemann, S., Sereda, M., Rossner, M., Stewart, H.,<br />
Suter, U., Meinck, H.-M., Griffiths, I.R., and Nave,<br />
K.-A. (1999). The ‚CMT rat‘: Peripheral neuropathy<br />
and dysmyelination caused by transgenic overexpression<br />
of PMP22. Annals N.Y. Acad. Sci. 883,<br />
254-261.<br />
Niemann, S., Sereda, M.W., Suter, U., Griffiths,<br />
I.R., and Nave, K.-A. (<strong>2000</strong>). Uncoupling of myelin<br />
assembly and Schwann cell differentiation by transgenic<br />
overexpression of PMP22. J. Neuroscience, 20,<br />
4120-4128.<br />
Schwab, M., Druffel-Augustin, S., Gass, P., Jung,<br />
M., Klugmann, M., Bartholomae, A., Rossner, M.,<br />
and Nave, K.-A. (1998). Neuronal basic helix-loophelix<br />
proteins (NEX, neuroD, NDRF): spatio-temporal<br />
expression and targeted disruption of the NEX<br />
gene in transgenic mice. J. Neurosci. 18, 1408-1418.<br />
Schwab, M.H., Bartholomä, A., Heimrich, B., Feldmeyer,<br />
D., Druffel-Augustin, S., Goebbels, S., Naya,<br />
F.J., Frotscher, M., Tsai, M.-J., and Nave, K.-A.<br />
(<strong>2000</strong>). Neuronal bHLH proteins (NEX and BETA/<br />
2NeuroD) regulate terminal granule cell differentiation<br />
in the hippocampus. J. Neurosci. 20, 3714-3724.<br />
Suter, U. and Nave, K.-A. (1999). Transgenic mouse<br />
models of CMT1A and HNPP. Annals N.Y. Acad. Sci.<br />
883, 247-253.<br />
Vouyiouklis, D.A., Werner, H., Griffiths, I.R., Stew-<br />
art, G.J., Nave, K.-A., and Thomson, C.E. (1998).<br />
Molecular cloning and transfection studies of M6b-2, a<br />
novel splice variant of a member of the PLP-DM20/M6<br />
gene family. J. Neurosci. Res. 52, 633-640.<br />
Werner, H., Jung, M., Klugmann, M., Sereda, M.,<br />
Griffiths, I., and Nave, K.-A. (1998). Mouse models<br />
of myelin diseases. Brain Pathol. 8, 771-793.<br />
THESES<br />
Staatsexamen<br />
Kassmann, C. (1999). Funktionelle Charakterisierung<br />
von basischen Helix-Loop-Helix Proteinen.<br />
Dissertations<br />
Rossner, M. (1998). <strong>Molekulare</strong> unf funktionale Charakterisierung<br />
zweier neuer Transkriptionsfaktoren,<br />
SHARP-1 und -2: Eine Rolle <strong>für</strong> bHLH Proteine in<br />
neuronaler Plastizität.<br />
Schwab, M. (1998). Herstellung und Analyse einer<br />
Mausmutante mit einer Nullmutation des NEX Gens.<br />
Klugmann, M. (1999). Funktionsanalyse des Myelin<br />
Proteolipid Proteins (PLP) und seines neuronalen<br />
Homologs M6A in vivo.<br />
Sereda, M. (1999). Ein transgenes Rattenmodell <strong>für</strong><br />
die Charcot-Marie-Tooth Erkrankung.<br />
95
STRUCTURE OF THE GROUP<br />
E-mail: nave@em.mpg.de<br />
Group lea<strong>der</strong> Nave, Klaus-Armin, Ph.D., Prof.<br />
Postdoctoral<br />
fellows Bartholomä, Angelika, Dr. rer. nat.<br />
Klugmann, Matthias, Dr. rer. nat.*<br />
Krämer, Evi, Dr. rer. nat.<br />
Niemann, Stefan, Dr. med.*<br />
Rossner, Moritz, Dr. rer. nat.*<br />
Schwab, Markus, Dr. rer. nat.*<br />
Sereda, Michael, Dr. med.*<br />
Yonemasu, Tomoko, Ph.D.*<br />
Ph.D. students Dimou. Leda, Dipl.-Biol.<br />
Goebbels, Sandra, Dipl.-Biol.<br />
Jung, Martin, Dipl.-Biol.<br />
Lappe, Corinna, Dipl. Biol.*<br />
Schardt, Anke, Dipl. Biol.*<br />
Werner, Hauke, Dipl. Biol.<br />
Technical<br />
assistants Druffel-Augustin, Silke<br />
Krischke, Hartmut<br />
* part of the time reported<br />
96<br />
Renato Paro<br />
Chromatin-Controlled Epigenetic Regulation<br />
of Transcription<br />
During the development of an organism, mechanisms<br />
of pattern formation program cells for particular functions<br />
and structures. The program code is based on<br />
a cell-specific differential repression or activation of<br />
master control genes. Once established such codes<br />
need to be maintained over many cell divisions during<br />
the growth phases and eventually also into adulthood,<br />
a process termed “cellular memory”. In this context,<br />
the stable and mitotically heritable inactivation of<br />
transcription (Silencing) becomes an important developmental<br />
function, necessary to generate and to maintain<br />
defined gene expression patterns in determined<br />
cells. The proteins of the Polycomb group (PcG) control<br />
the permanent inactivation of program coding factors<br />
like the HOX genes. PcG proteins appear to inactivate<br />
their target genes by generating heterochromatin-like<br />
structures. Conversely, the proteins of the trithorax<br />
group (trxG) counteract the silencing role of<br />
the PcG and maintain the active expression state of the<br />
target genes. Thus, the maintenance of gene expression<br />
patterns is controlled at the levels of higher or<strong>der</strong><br />
chromatin structures. Chromatin components such as<br />
histones, through their modifications, and other chromatin<br />
proteins seem to be able to generate an “epigenetic<br />
code” that influences the processing of the un<strong>der</strong>lying<br />
genetic information. We are studying and trying<br />
to decipher this epigenetic code in the fruit fly Drosophila<br />
melanogaster by assessing the molecular functions<br />
of the PcG and trxG proteins in the chromatinbased<br />
control of HOX gene expression.<br />
Chromatin-based silencing mechanisms appear to have<br />
many evolutionary conserved features. Thus, their<br />
analysis in a genetically well tractable system like<br />
Drosophila serves as a paradigm for other epigenetic<br />
phenomena like the mammalian X-chromosome inactivation<br />
or genomic imprinting. In addition, there is<br />
increasing evidence that the missregulation of elements<br />
of cellular memory in adulthood can result<br />
in tumorigenesis in mammals. We would like to<br />
un<strong>der</strong>stand and to identify the common denominator<br />
involved in these apparently diverse processes. What<br />
creates and forms particular chromatin structures that<br />
maintain defined gene expression patterns? Is the<br />
DNA-encoded genetic information overlaid by an epigenetic<br />
code, and how is this code transmitted from<br />
one cell generation to the next?<br />
I. Chromosomal elements conferring epigenetic<br />
inheritance<br />
G. Cavalli; S. Ehrhardt; F. Lyko; M. Prestel; DFG,<br />
HFSP (in collaboration with A. Surani (Wellcome,<br />
Cambridge) and R. Ohlson (Univ. of Uppsala))<br />
Using chromatin-IP (X-CHIP) methodology developed<br />
in the lab, we have identified several chromosomal<br />
elements of the bithorax complex (BX-C)<br />
through which the PcG and trxG proteins exert their<br />
functions. We have shown in a particular transgenic<br />
set-up that chromosomal elements, such as Fab-7 controlling<br />
the Abdominal-B gene in the BX-C, are able<br />
to maintain a decision made early during embryogenesis<br />
through many mitotic cell divisions during development.<br />
Fab-7 is switched from a silenced (PcGcontrolled)<br />
to an activated (trxG-controlled) chromatin<br />
state by a pulse of embryonic transcription. Once<br />
activated the expression of a nearby reporter gene<br />
becomes independent of the primary transcription<br />
factor. Surprisingly, in the transgenic system, an active<br />
97
Fab-7 state can also be transmitted meiotically in a<br />
substantial portion of the progeny (Figure 1).<br />
While we initially established the system using Fab-7,<br />
we have now demonstrated a similar maintenance<br />
function for two other elements of the BX-C, MCP<br />
and BXD. Thus, our results indicate that in genetic<br />
complexes containing master control genes like the<br />
Figure 1. In a transgene construct an activated CMM is<br />
stable during mitotic cell divisions and to a certain degree<br />
also stable through meiosis. This result shows that in principle<br />
it is possible to transmit a chromatin-based epigenetic<br />
state through the germ line into the next generation.<br />
98<br />
homeotic genes, defined chromosomal elements act<br />
as “cellular memory modules” (CMMs) capable of<br />
memorizing active or repressed gene expression patterns<br />
over developmental time.<br />
In the previous period, we have used Drosophila to<br />
identify chromosomal elements controlling mammalian<br />
epigenetic mechanisms like genomic imprinting.<br />
In collaboration with the Surani lab we have identified<br />
an upstream element of the imprinted H19 gene acting<br />
as a silencer in Drosophila. The subsequent deletion<br />
of this element in the endogenous murine H19 locus<br />
by the Surani lab resulted in the loss of the imprint. In<br />
parallel, we could demonstrate in collaboration with<br />
the Ohlson lab that the nucleosomal patterning of the<br />
upstream region containing the imprinting element of<br />
H19 in transgenic flies highly resembles the endogenous<br />
mouse H19 pattern. Thus, conserved features<br />
seem to be involved in maintaining the epigenetically<br />
controlled imprinted states of H19. Indeed, in another<br />
approach we were able to demonstrate that a different<br />
chromosomal element, necessary for the maintenance<br />
of H19 expression, in transgenic Drosophila is bound<br />
and controlled by a member of the PcG (PSC). These<br />
results witness the usefulness of Drosophila in studying<br />
basic conserved mechanisms of chromatin regulation<br />
in epigenetic processes.<br />
II. Epigenetic marking of CMMs<br />
B. Breiling, G. Cavalli, C. Maurange, G. Rank,<br />
DFG, HFSP (in collaboration with P. Becker (Univ.<br />
of Munich))<br />
PcG proteins show an extended binding profile at<br />
CMMs. At the molecular level we could demonstrate<br />
a highly specific binding of PC protein through its<br />
C-terminal part with in vitro reconstituted nucleosomes.<br />
Surprisingly, we found that also in the acti-<br />
vated state PcG proteins are bound to the CMM. Conversely,<br />
trxG proteins are still bound to a repressed<br />
CMM. Thus, the interplay between PcG and trxG<br />
members appears to be vital for the proper function<br />
of CMMs. What defines a CMM, like Fab-7, to be in<br />
an active or in an inactive mode and how this tag<br />
is maintained over many cell generations, are questions<br />
we are currently addressing. We could demonstrate<br />
that hyperacetylated histone H4 tags an activated<br />
CMM, and this epigenetic mark is transmissible<br />
mitotically (Figure 2).<br />
We are now assessing how CMMs become switched<br />
from an apparent silenced default state to an activated<br />
state. After embryogenesis the epigenetic state<br />
of CMMs become fixed and cannot be changed anymore.<br />
Thus, a specific event occurring during this<br />
early stage appears to switch a CMM. After passing a<br />
specific developmental threshold, the epigenetic state<br />
is fixed and clonally inherited in subsequent cell generations.<br />
We have evidence that early transcription<br />
through the element might be important to change<br />
the overall chromatin structure and thus the epigenetic<br />
state of the CMM. Additionally, we are testing<br />
the extent histone modifications at CMMs potentially<br />
have as primary epigenetic tags and how such modifications<br />
can be inherited at DNA replication and at<br />
mitosis.<br />
III. The TRITHORAX protein in cancer formation<br />
I. Chen-Muyrers, S. Schönfel<strong>der</strong>, DKFZ-MOS,<br />
DFG (in collaboration with E. Canaani (Weizmann,<br />
Rehovot) and V. Orlando (DIBIT, Milano))<br />
The gene trithorax (trx), the name giving member of<br />
the trxG, was identified by its requirement for normal<br />
expression of multiple homeotic genes of the bithorax<br />
and Antennapedia complex. Different homeotic<br />
genes and their promoters are affected differently in<br />
trx mutants, suggesting a coordinated spatial and tem-<br />
Figure 2. Immunostaining of larval<br />
polytene chromosomes at the<br />
integration site of a Fab-7 (CMM)<br />
transgene construct. After embryonic<br />
activation the Fab-7 construct<br />
remains marked by histone<br />
H4 hyperacetylation also at larval<br />
stages (pannel D; arrow). Control<br />
constructs or non-activated Fab-7<br />
do not show a H4 hyperactylation<br />
signal (pannels A-C) nor is H3<br />
hyperacetylation observed in any<br />
of the cases (pannels F-H).<br />
99
poral requirement for trx in or<strong>der</strong> to achieve normal<br />
expression patterns. We have shown with X-CHIP a<br />
dynamic association of TRX with CMMs and promoters<br />
of the BX-C and found a close cooperation of TRX<br />
with PcG members at CMMs.<br />
Several members of the Drosophila trxG have counterparts<br />
in humans and mice. The mixed-lineage leukaemia<br />
(MLL) gene (also known as ALL-1, HRX), a<br />
TRX homologue, was found through its implication in<br />
the majority of infantile acute lymphocytic and mixed<br />
lineage leukemias. Disruption of the murine Mll gene<br />
by gene targeting causes homeotic transformations of<br />
the vertebrae in heterozygotes and loss of homeotic<br />
gene expression, supporting the notion that Mll is a<br />
functional equivalent of trx.<br />
To date, not much is known at the molecular level<br />
how translocation of MLL with various other chromosomal<br />
locations generate leukemia. At least 23 fusion<br />
MLL proteins involved in leukemia have been identified.<br />
As part of our collaborative efforts with<br />
the Canaani lab, we have generated transgenic flies<br />
expressing full length MLL, and fusion proteins<br />
MLL-AF9, MLL-AF4, and MLL-AF17 un<strong>der</strong> GAL4<br />
control. Expression of these proteins in Drosophila<br />
may reveal their effects by determining with which<br />
pathway in Drosophila development they influence.<br />
Crosses between our transgenic flies and various<br />
GAL4 drivers revealed that only MLL-AF9 and<br />
MLL-AF17 constructs had a slight effect on Drosophila<br />
development. When we took a closer look at the<br />
molecular level, we could not detect the proteins on<br />
Western, but at the RNA level, we see transcripts from<br />
all four constructs. Interestingly, this same effect is<br />
also observed with MLL-AF9 in mouse. We are currently<br />
testing whether overexpression of the murine<br />
proteins results in cell death as recent published observations<br />
link MLL with apoptosis.<br />
100<br />
IV. Drosophila as a model system to identify<br />
components involved in the processing<br />
and the function of the human Amyloid Precursor<br />
Protein<br />
G. Merdes, B. Brückner, DFG (in collaboration<br />
with K. Beyreuther (<strong>ZMBH</strong>))<br />
The human amyloid precursor protein (APP) is an<br />
integral membrane protein with two homologues in<br />
invertebrates: the amyloid protein-like protein 1 (C.<br />
elegans, APL-1) and the amyloid precursor proteinlike<br />
protein (D. melanogaster, APPL). Flies deficient<br />
for expression of APPL show a phototaxis impairment<br />
that can be rescued by the expression of human APP<br />
suggesting an evolutionary conservation of APP function.<br />
The importance of APP in the pathogenesis of<br />
Alzheimer’s disease (AD) became apparent through<br />
the identification of distinct mutations in the gene<br />
causing early onset familial AD. The major component<br />
of the plaques and amyloid found in senile Alzheimer<br />
patients is a proteolytic product of APP, the<br />
βA4 peptide. APP is cleaved in vivo by several proteases<br />
termed α−, β−, and γ−secretase. Cleavage of<br />
APP by the α−secretase prevents the production of<br />
βA4. Despite the availability of APP-null mutants and<br />
transgenic mice expressing human APP, the physiological<br />
role of APP remains unknown and many factors<br />
involved in the processing of APP have not yet<br />
been clearly identified. Therefore we have established<br />
Drosophila melanogaster as a model system to study<br />
the function and processing of APP.<br />
Various forms of human APP were expressed in<br />
Drosophila tissue culture cells as well as in transgenic<br />
fly lines. Studies in both systems revealed that also in<br />
flies APP is processed in a similar way as in mammalian<br />
cells in regard to the α- and γ-cleavages. In addition,<br />
transgenic flies expressing full-length forms of<br />
APP in the wing imaginal discs are characterized by a<br />
blistered-wing phenotype, suggesting that the expression<br />
of human APP interferes with the cell adhesion<br />
between the dorsal and ventral epithelial cell monolayers<br />
of the wing (Figure 3). To define the involvement<br />
of APP in the blistered wing phenotype in more<br />
detail as well as to identify factors involved in the processing<br />
of APP, we perform biochemical and genetic<br />
studies. Since the observed wing phenotype depends<br />
on full-length APP, an increased processing of APP,<br />
e.g. by the simultaneous overexpression of an APPspecific<br />
protease, results in a suppression of the phenotype.<br />
This observation enables us to identify proteins<br />
involved in the processing of APP by the use of<br />
genetic screens and to subsequently identify the corresponding<br />
mammalian homologues. In a similar way,<br />
gene products required for the effect of APP on cell<br />
adhesion and therefore for APP function, can also be<br />
identified by genetic screens. Mutations in these genes<br />
result in suppression or an enhancement of the blis-<br />
tered wing phenotype, dependent on their function.<br />
Drosophila can be mutagenized by the use of chemicals<br />
or by the use of the transposable P-element.<br />
The advantage of P-element mutagenesis is the easier<br />
identification of the mutagenized genes by isolating<br />
and sequencing the flanking genomic DNA. Therefore<br />
we have recently performed a P-element based genetic<br />
screen as well as screened collections of already existing<br />
Drosophila mutants. The analysis of 150 000 flies<br />
with potential new P-element insertion sites resulted<br />
in the isolation of several enhancer- and suppressormutations,<br />
together with several genes interfering with<br />
wing patterning. The corresponding genes, which we<br />
are currently characterizing further, encode factors<br />
involved in signal transduction, maintenance of cell<br />
shape and cell adhesion, and protein processing.<br />
We utilize the fruit fly Drosophila melanogaster as<br />
a genetic model system. Since Thomas Hunt Morgan<br />
introduced Drosophila as a model organism into genetics<br />
at the beginning of this century and isolated the<br />
Figure 3. Expression of different<br />
isoforms of human APP in transgenic<br />
Drosophila. Only full-length<br />
APP result in a blistered wing<br />
phenotype.<br />
101
first mutation, thousands of genes and mutations have<br />
been isolated and characterized, making Drosophila<br />
one of the best-studied multicellular organisms. In<br />
Drosophila many factors and basic principles of<br />
development and differentiation have been uncovered,<br />
which were subsequently found to also play important<br />
roles during the development of mammals, e.g. morphogens<br />
and the homeodomain. Surprisingly, despite<br />
the difference in morphology, even gene products<br />
required for the development of specific organs have<br />
been evolutionary conserved in structure and function.<br />
This conservation of fundamental developmental and<br />
cellular principles as well as the vast assortment of<br />
genetic, cell and molecular biological tools available,<br />
allows us to study biological problems of medical relevance<br />
in Drosophila and to identify conserved factors<br />
and mechanisms required for epigenetics.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(Sachbeihilfe and Graduiertenkolleg “Signalsysteme<br />
und Genexpression in entwicklungsbiologischen<br />
Modellsystemen”, from the Human Frontier<br />
Science Program, from the Deutsche-Israelische<br />
Zusammenarbeit in <strong>der</strong> Krebsforschung (DKFZ/<br />
MOS), from the BMBF (Humangenomprojekt) and<br />
the Fonds <strong>der</strong> Chemischen Industrie.<br />
PUBLICATIONS<br />
Brenton, J.D., Ainscough, J.F., Lyko, F., Paro, R., and<br />
Surani, M.A. (1998). Imprinting and gene silencing<br />
in mice and Drosophila. Novartis Found Symp. 214,<br />
233-442.<br />
Cavalli, G. and Paro, R. (1998). Chromo-domain proteins:<br />
linking chromatin structure to epigenetic regula-<br />
102<br />
tion. Curr. Opin. Cell. Biol. 10, 354-360.<br />
Cavalli, G. and Paro, R. (1998). The Drosophila Fab-7<br />
chromosomal element conveys epigenetic inheritance<br />
during mitosis and meiosis. Cell 93, 505-518.<br />
Fossgreen, A., Brückner, B., Czech, C., Masters,<br />
C.L., Beyreuther, K., and Paro, R. (1998). Transgenic<br />
Drosophila expressing human amyloid precursor protein<br />
show gamma-secretase activity and a blisteredwing<br />
phenotype. Proc. Natl. Acad. Sci. U.S.A. 95,<br />
13703-13708.<br />
Gasser, S.M., Paro, R., Stewart, F., and Aasland, R.<br />
(1998). The genetics of epigenetics. Cell. Mol. Life<br />
Sci. 54, 1-5.<br />
Lyko, F., Buiting, K., Horsthemke, B., and Paro, R.<br />
(1998). Identification of a silencing element in the<br />
human 15q11-q13 imprinting center by using transgenic<br />
Drosophila. Proc. Natl. Acad. Sci. U.S.A. 95,<br />
1698-1702.<br />
Orlando, V., Jane, E.P., Chinwalla, V., Harte, P.J., and<br />
Paro, R. (1998). Binding of trithorax and Polycomb<br />
proteins to the bithorax complex: dynamic changes<br />
during early Drosophila embryogenesis. EMBO J.<br />
17, 5141-5150.<br />
Paro, R., Strutt, H., and Cavalli, G. (1998). Heritable<br />
chromatin states induced by the Polycomb and trithorax<br />
group genes. Novartis Found. Symp. 214, 51-61.<br />
Paro, R. (1998). Das Zellgedächtnis. Biospektrum 6,<br />
21-24.<br />
Breiling, A., Bonte, E., Ferrari, S., Becker, P.B., and<br />
Paro, R. (1999). The Drosophila Polycomb protein<br />
interacts with nucleosomal core particles in vitro via its<br />
repression domain. Mol. Cell. Biol. 19, 8451-8460.<br />
Cavalli, G. and Paro, R. (1999). Epigenetic inher–<br />
itance of active chromatin after removal of the main<br />
transactivator. Science 286, 955-958.<br />
Dietzel, S., Niemann, H., Brückner, B., Maurange, C.,<br />
and Paro, R. (1999). The nuclear distribution of Polycomb<br />
during Drosophila melanogaster development<br />
shown with a GFP fusion protein. Chromosoma 108,<br />
83-94.<br />
Lyko, F. and Paro, R. (1999). Chromosomal elements<br />
conferring epigenetic inheritance. Bioessays 21,<br />
824-832.<br />
Paro, R. (1999) Trithorax Group Genes. In Encyclopedia<br />
of Molecular Biology (ed. Creighton, T.E.) Wiley,<br />
New York, pp 2670-2672.<br />
Paro, R. (<strong>2000</strong>). Mapping Protein Distributions on<br />
Polytene Chromosomes by Immunostaining. In Drosophila<br />
Protocols Manual (ed. Ashburner, M.) CSHL<br />
Press, CHS, (in press)<br />
THESES<br />
Diploma<br />
Erhardt, Sylvia (1998). Etablierung und Charakterisierung<br />
transgener Drosophila melanogaster mit<br />
einem cis-regulatorischen Element geprägter Gene <strong>der</strong><br />
Maus und die Analyse <strong>der</strong> am Gen-Silencing beteiligten<br />
Faktoren.<br />
Dissertations<br />
Breiling, Achim (1998). Charakterisierung <strong>der</strong> funk-<br />
tionellen Aufgaben <strong>der</strong> beiden evolutionär konser–<br />
vierten Bereiche des Polycomb-Proteins aus Drosophila<br />
melanogaster: die Chromodomäne und <strong>der</strong><br />
C-Terminus.<br />
Lyko, Frank (1998). Identifikation und Charakterisie–<br />
rung von Silencing-Elementen in cis-regulatorischen<br />
Regionen geprägter Säugergene mit Hilfe transgener<br />
Drosophila melanogaster.<br />
(GFM-Preis <strong>für</strong> beste Doktorarbeit)<br />
STRUCTURE OF THE GROUP<br />
E-mail: paro@sun0.urz.uni-heidelberg.de<br />
Group lea<strong>der</strong> Paro, Renato, Prof. Dr.<br />
Research associates Cavalli, Giacomo, Dr.*<br />
Merdes, Gunter, Dr.<br />
Ph.D. students Breiling, Achim, Dipl. Biol.*<br />
Brückner, Bodo, Dipl. Biol.<br />
Chen-Muyrers, Inhua<br />
Lyko, Frank, Dipl. Biol.*<br />
Maurange, Cédric*<br />
Prestel, Matthias, Dipl. Biol.*<br />
Rank, Gerhard, Dipl. Biol.<br />
Diploma students Erhardt, Sylvia*<br />
Schönfel<strong>der</strong>, Stefan*<br />
Techn. assistants Albrecht, Steffen *<br />
Bas-Orth, Carlos *<br />
Friedrich, Daniela *<br />
Ehret, Heidi<br />
Eysel, Georg *<br />
Mund, Cora *<br />
Schwarz, Gabriele<br />
*part of the time reported<br />
103
Frank Sauer<br />
Mechanisms of Transcriptional Regulation<br />
Our research focuses on the molecular mechanisms<br />
governing regulation of eukaryotic gene expression.<br />
The capability of living organisms to control gene<br />
expression is essential for the execution of complex<br />
biological events, such as body pattern formation or<br />
cell differentiation, as well as the response to extra-<br />
and intra-organismal stimuli. Eukaryots have evolved<br />
refined mechanisms that regulate the precise temporal<br />
and spatial expression of genes. At the molecular<br />
level, eukaryotic gene expression is predominantly<br />
regulated at the level of transcriptional initiation. A<br />
widely accepted model proposes that initiation of<br />
transcription requires the precisely regulated assembly<br />
of distinct multi-protein complexes on eukaryotic<br />
genes. Major efforts have led to the identification of<br />
the key players that initiate m-RNA synthesis. The<br />
first player is the eukaryotic gene, that in general<br />
is comprised of a protein coding region, which<br />
is flanked by two essential cis-regulatory regions:<br />
core-promoter and enhancer/silencer. The core-promoter<br />
defines the start-site of transcription while the<br />
enhancer/silencer mediate the precise temporal and<br />
spatial expression of the coding region. The core promoter<br />
is the target for the general RNA polymerase II<br />
(pol II) transcription machinery (GTM). This multiprotein<br />
complex contains general transcription factors<br />
(TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH), RNA<br />
polymerase II, the catalytic motor of m-RNA synthesis<br />
and associated protein-complexes. Enhancer/<br />
silencer-regions are characterized by their repertoire<br />
of DNA-sequences, which serve as targets for transcription<br />
factors, a diverse family of DNA-binding<br />
Figure 1. Dorsal-dependent transcriptional activation is alleviated in TAF250-mutant embryos. In situ hybridization of<br />
whole mount Drosophila embryos (left and middle panel) showing twist and snail expression. Note that the expression of<br />
both twist and snail is significantly reduced in TAF250 mutant embryos. (Right panel) Cuticula preparations of Drosophila<br />
embryos. Note that the Drosophila body pattern is significantly altered in TAF250-mutant embryos.<br />
104<br />
proteins. It is thought, that transcription factors contact<br />
components within the general transcription machinery<br />
to either activate or repress the process of transcriptional<br />
initiation. This communication between<br />
transcription factors and GTM requires in many cases<br />
so called co-factors, which function as receivers of the<br />
regulatory signals provided by transcription factors.<br />
Although co-factors are a diverse group of proteins, a<br />
common feature of these proteins is their direct interaction<br />
with transcription factors and recruitment of<br />
co-factors has been established as an essential step<br />
of transcriptional regulation. It is thought that co-factors<br />
function as molecular bridge between transcription<br />
factors and GTM. The finding that the same cofactors,<br />
which function as molecular bridge between<br />
transcription factors and GTM, can mediate posttranslational<br />
modification of histones has added an unexpected<br />
level of complexity to the model of transcriptional<br />
regulation. Histones (H1, H2A, H2B, H3,<br />
H4) are the basic building block of nucleosomes, the<br />
most fundamental structural entity within chromatin.<br />
Nucleosomes inhibit the most fundamental step of<br />
transcriptional regulation, namely the interaction of<br />
transcription factors and GTM with DNA. To overcome<br />
this “nucleosome-barrier” it is thought that transcription<br />
factors recruit co-factors, which by posttranslational<br />
modification of histones establish transcriptional<br />
active or inactive chromatin structures.<br />
Abundant evidence has been provided that co-factor<br />
mediated acetylation of core-histones is correlated<br />
with transcriptional activation while deacetylation is<br />
concomitant with repression. Fuelled by the functional<br />
diversity of co-factors deciphering the molecular<br />
mechanisms for how co-factors convert regulatory<br />
signals into activated or repressed states of transcription<br />
lies at the heart of the transcription field.<br />
TAFs mediate transcriptional activation in<br />
Drosophila<br />
Anh-Dung Pham<br />
Transcription factors contact one or multiple components<br />
of the GTM to activate transcription. One target<br />
of activators is the general transcription factor TFIID<br />
a multi-protein complex comprised of the TATA-box<br />
binding protein (TBP) and at least eight TBP-associated<br />
factors (TAF II s). Distinct TAF II s serve as targets<br />
for different activation domains. As TAF II -activator<br />
interactions mediate transcriptional activation in<br />
vitro, TAF II s are thought to function as co-activators,<br />
receivers, of activation signals. However, the role and<br />
function of TAF II s for transcriptional activation in<br />
vivo remains unclear. We have used Drosophila as a<br />
model system to investigate the functional importance<br />
of TAF II s for activation of transcription. To identify<br />
transcription factors, which activate transcription in a<br />
TAF II -dependent manner, we made use of TAF II 250<br />
mutant flies. As TAF II 250 is the central subunit in the<br />
TBP-TAF-complex, a disruption of this TFIID-subunit<br />
is thought to abrogate the structure and function<br />
of TFIID and, hence TFIID-dependent transcriptional<br />
activation. We could show that a mutation within<br />
TAF II 250 alleviates Dorsal-dependent transcriptional<br />
activation (Figure 1). The Rel transcription factor<br />
Dorsal establishes dorso-ventral polarity in the early<br />
Drosophila embryo by activating the expression of<br />
different tissue-determining genes in ventral and lateral<br />
cells. We could show that Dorsal interacts with<br />
two TAF II -subunits, TAF II 110 and TAF II 60. By using a<br />
reconstituted Drosophila in vitro transcription system<br />
and in vitro assembled partial TBP-TAF II -complexes<br />
we could show that interactions between Dorsal and<br />
TAF II 110 and/or TAF II 60 mediate transcriptional activation<br />
in vitro. To provide evidence for the biological<br />
relevance of these Dorsal: TAF II -interactions in vivo,<br />
105
we made use of TAF mutant flies. The expression<br />
of Dorsal target genes was significantly reduced in<br />
TAF110 and/or TAF60 mutant embryos indicating<br />
that these co-activators contribute to Dorsal-dependent<br />
transcriptional activation in Drosophila. Our<br />
results provide evidence that TAF II s play an important<br />
role for transcriptional activation in Drosophila.<br />
Figure 2. Model for Dorsal-dependent transcriptional activation.<br />
For details see ”TAFs mediate transcriptional activation<br />
in Drosophila”.<br />
Mechanisms un<strong>der</strong>lying STAT-dependent transcriptional<br />
regulation in Drosophila<br />
Simona Kwocynski<br />
STATs (signal transducers and activators of transcription)<br />
are phylogenetic highly conserved latent cytoplasmic<br />
transcription factors that have been identified<br />
in Dictyostelium, Drosophila, mouse and human.<br />
STATs activate the expression of specific target genes,<br />
which encode proteins that are involved in complex<br />
biological processes such as immune response or<br />
embryogenesis. Although the biological function and<br />
importance of STATs has well established, the molec-<br />
106<br />
ular mechanisms un<strong>der</strong>lying STAT-dependent transcriptional<br />
activation remain poorly un<strong>der</strong>stood. To<br />
address this question we have established a STATdependent<br />
in vitro transcription assay that enables us<br />
to characterize the co-factor requirements for STATmediated<br />
transcriptional activation. The functional<br />
relevance of these co-factors has been tested using<br />
reconstituted in vitro transcription systems supplemented<br />
with mutant co-factors. We use the Drosophila-embryo<br />
as model system to determine the biological<br />
relevance of the in vitro identified co-factors for<br />
STAT-dependent transcriptional activation.<br />
Mechanisms of transcriptional repression in<br />
Drosophila<br />
Thorsten Belz<br />
The molecular mechanisms un<strong>der</strong>lying repression of<br />
transcription in eukaryotic cells remain opaque. We<br />
use the Drosophila proteins Snail and Krüppel as<br />
models to decipher modes of transcriptional repression<br />
in Drosophila. The gap gene krüppel (Kr) is<br />
expressed in the central region of the blasto<strong>der</strong>m<br />
Drosophila embryo and encodes a zinc-finger type<br />
transcription factor (Krüppel). Genetic analyses have<br />
established that Krüppel represses the expression of<br />
the gap-gene giant and the second expression domain<br />
of the pair-rule gene even-skipped (eve). Snail (sna)<br />
is expressed in 18 ventral-most cells of the embryo.<br />
Sna encodes a zinc-finger type transcription factor<br />
that represses the expression of neuroecto<strong>der</strong>m-specific<br />
genes, including rhomboid (rho) and singleminded<br />
(sim). We use a combination of biochemistry<br />
and genetics to decipher the molecular mechanisms<br />
un<strong>der</strong>lying Snail- and Krüppel-dependent transcriptional<br />
repression.<br />
Identification of histone modifying enzymes<br />
Christian Beisel<br />
Various posttranslational modifications of histones<br />
have been described. Although these modifications<br />
have been correlated with transcriptional activation,<br />
their precise role for transcriptional regulation remains<br />
unknown. Two of the least un<strong>der</strong>stood histone modifications<br />
are methylation and ubiquitination of histones.<br />
The predominant histones, which are methylated,<br />
are H3 and H4. Different studies have linked<br />
histone methylation with regulation of gene expression.<br />
In Drosophila changes in the gene expression<br />
pattern in response to heat shock are concomitant<br />
with changes in the methylation pattern of histones.<br />
In Tetrahymena, transcriptionally active macronuclei<br />
contain histone methyltransferase (HMT)-activity<br />
whereas transcriptional inactive micronuclei do not.<br />
It, therefore, has been proposed that histone methylation<br />
plays a role in the reconfiguration of chromatin<br />
un<strong>der</strong> stress conditions. A functional link between histone<br />
methylation and transcriptional regulation came<br />
with the finding that the co-activator CARM-1 possesses<br />
a histone-specific HMT-activity. Like methylation,<br />
ubiquitination of histones has been correlated<br />
with the execution of various DNA-dependent processes.<br />
In Tetrahymena, ubiquitinated histones are<br />
present in transcriptional active chromatin structures<br />
implying that ubiquitination of histones correlates<br />
with activation of transcription.<br />
However, until now the role and function(s) of histone-methylation<br />
and -ubiquitination for transcriptional<br />
activation and the responsible enzymes remain<br />
unknown. It is our aim to identify the enzyme(s)<br />
responsible for methylation and ubiquitination of<br />
histones.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (Graduiertenkolleg “Signalsysteme und Genexpression<br />
in entwicklungsbiologischen Modellsystemen”,<br />
Projekt-’Sachbeihilfen’) and the BMBF.<br />
PUBLICATIONS<br />
Schock, F., Sauer, F., Jäckle., H and Purnell, BA.<br />
(1999). Drosophila head segmentation factor buttonhead<br />
interacts with the same TATA box-binding protein-associated<br />
factors and in vivo DNA targets as<br />
human Sp1 but executes a different biological program.<br />
Proc. Natl. Acad. Sci. USA 96, 5061-5065.<br />
Pham, A.D., Müller, S. and Sauer F. (1999). Meso<strong>der</strong>m-determining<br />
transcription in Drosophila is alleviated<br />
by mutations in TAF II 60 and TAF II 110. Mech.<br />
Dev. 84, 3-16.<br />
La Rosee-Borggreve, A., Ha<strong>der</strong>,T., Wainwright, D.,<br />
Sauer F. and Jäckle H. (1999). Hairy stripe 7 element<br />
mediates activation and repression in response to different<br />
domains and levels of Krüppel in the Drosophila<br />
embryo. Mech. Dev. 89, 133-140.<br />
Pham, A.-D., and Sauer, F. (<strong>2000</strong>). Ubiquitinactivating/conjugating<br />
activity of TAF II 250 mediates<br />
activation of gene expression in Drosophila. Science<br />
(in press).<br />
107
THESES<br />
Diploma<br />
Belz, Thorsten (1998). Untersuchungen zur Assoziation<br />
von Repressoren <strong>der</strong> Transkription aus Drosophila<br />
melanogaster mit Histon-Deacetylase-Aktivität.<br />
Kwoczynski, Simona (1998). Genetische Untersu–<br />
chung zum Mechanismus <strong>der</strong> STAT-abhängigen Transkriptionsaktivierung<br />
in Drosophila melanogaster.<br />
Bierlein, Nicole (1999). Untersuchungen zum Einfluß<br />
von Mutationen des TBP-assoziierten Faktors<br />
TAF II 250 auf die differentielle Genexpression in<br />
Drosophila melanogaster.<br />
STRUCTURE OF THE GROUP<br />
E-mail: f. sauer@mail.zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Sauer, Frank, Dr.<br />
Ph.D. students Beisel, Christian<br />
Belz, Thorsten<br />
Kwoczynski, Simona<br />
Dung-Pham, Anh<br />
Diploma students Egle, Markus*<br />
Bogin, Jochen*<br />
Tech. assistants Leibfried, Ute*<br />
Müller, Sandra<br />
* part of the time reported<br />
108<br />
Hans Ulrich Schairer<br />
I. Stigmatella aurantiaca, a Prokaryotic<br />
Organism for Studying Intercellular Signalling<br />
and Morphogenesis<br />
The organisms<br />
Stigmatella aurantiaca belongs to the myxobacteria<br />
that are Gram-negative soil bacteria. Myxobacteria<br />
show both, features of unicellular and multicellular<br />
organisms. As the eukaryotic organism Dictyostelium<br />
dicoideum they are thought to lie on the boundary<br />
between unicellular and multicellular organisms.<br />
Myxobacteria grow and divide as separate cells. But<br />
they may be regarded as a multicellular organisms<br />
whose cells feed in swarms and which un<strong>der</strong> conditions<br />
of starvation, assemble to well defined regular<br />
three dimensional structures called fruiting bodies<br />
which enclose about 10 5 dormant cells, the myxospores.<br />
The shape of the fruiting bodies is species specific<br />
and is genetically determined. The fruiting body<br />
of S. aurantiaca consists of a stalk bearing several<br />
sporangioles on branches at its top.<br />
Myxobacteria secrete hydrolytic enzymes together<br />
with slime with which they degrade particulate organic<br />
matter of the soil. It has been shown that the growth<br />
rate increases with cell density if myxobacteria were<br />
grown on a macromolecular substrate as sole nutrient,<br />
such as casein. This suggests that cells feed co-operatively<br />
and the association in a swarm allows them to<br />
feed more efficiently. The advantage of cooperative<br />
feeding may have driven the evolution of fruiting body<br />
formation. When nutrients are again available after a<br />
period of starvation, myxospores germinate and form<br />
vegetative cells. The multicellular nature of the fruiting<br />
body ensures that a swarm of cells is formed for a<br />
new growth cycle.<br />
Myxobacteria move by gliding on solid surfaces. This<br />
facilitates the stabilisation of a swarm and of fruiting<br />
body formation. Gliding permits tight cell-cell contact<br />
and efficient signal exchange between the cells<br />
by diffusible molecules. Both features are a prerequisite<br />
for the transmission of positional information of<br />
the single cell necessary for the coordination of the<br />
metabolism and movement of the cell in the course<br />
of fruiting body formation. One of the developmental<br />
signals – Stigmolone – that is involved in early cell<br />
aggregation has been recently isolated and characterised<br />
by Wulf Plaga et al.<br />
Apart from their ability to form fruiting bodies, myxobacteria<br />
form a broad range of secondary metabolites.<br />
All these unique features are reflected in the size of the<br />
genome and its organisation. The size of the myxobacterial<br />
genome has been shown to be about 9.5 Mbp.<br />
A gene cluster involved in S. aurantiaca fruiting<br />
body formation<br />
Susanne Müller, Barbara Silakowski and Diana Hofmann<br />
To investigate the genes involved in S. aurantiaca<br />
fruiting body formation and the co-ordination of their<br />
expression, Tn5 transposon insertion mutagenesis was<br />
performed. Three different mutant types impaired in<br />
fruiting body formation were detected by screening<br />
the insertional mutants. They include mutants that<br />
form neither fruiting bodies nor aggregates. mutants<br />
that aggregate to unstructured clumps, and mutants<br />
that un<strong>der</strong>go only part of the differentiation process.<br />
One of the mutants (AP182) that formed clumps<br />
during starvation was analysed further. Mixing of the<br />
109
cells of this strain with those of a mutant (AP191),<br />
which was unable to form aggregates prior to starvation,<br />
lead to a partial phenotypic complementation.<br />
Instead of clumps, a mushroom-like structure, similar<br />
to a champignon was obtained. Sequencing of the<br />
mutant gene of strain AP182 and of the adjacent<br />
genomic segments resulted in four open reading<br />
frames that were involved in fruiting body formation.<br />
One of the genes, fbfB, and the other three genes,<br />
fbfA, fbfC, and fbfD are arranged in a divergent orientation.<br />
FbfB shows significant homology to the<br />
secreted copper enzyme galactose oxidase from the<br />
fungus Dactylium dendroides. fbfA encodes a polypeptide<br />
that is homologous to chitin synthases. The<br />
start codon of fbfC overlaps with the stop codon of<br />
fbfA. FbfC has no significant homology to any of the<br />
known proteins. FbfD has similarities to the human<br />
phosphoprotein synapsin I. Inactivation of either of<br />
these genes by insertion of the neo gene cassette<br />
resulted in mutants that formed only unstructured<br />
clumps during starvation. This indicates the gene products<br />
of the fbf-genes to be involved in fruiting body<br />
formation. No additional open reading frame involved<br />
in fruiting was detected downstream of fbfD. Mixing<br />
of cells from either the fbfA or the fbfB mutant with<br />
those of the non-aggregating strain AP191 before starvation<br />
resulted in mushroom-like fruiting bodies with<br />
the form of a morel or a champignon, respectively.<br />
These forms are observed during wild-type fruiting<br />
body formation 12 or 15 hours after the beginning of<br />
starvation.<br />
The partial phenotypic complementation suggests that<br />
factors involved in fruiting and which are lacking in<br />
one mutant may be obtained from the other. A reason<br />
for the incomplete phenotypic complementation may<br />
be that not all substances involved in fruiting (e.g.,<br />
intracellular macromolecules) which are lacking in<br />
110<br />
one of the strains can be supplemented by the other<br />
mutant and vice versa.<br />
For the analysis of the time dependence of the expression<br />
of the genes fbfA, fbfB, fbfC, and fbfD during<br />
fruiting body formation or indole induced sporulation<br />
merodiploid strains were constructed. They harboured<br />
the wild type genes and in addition a 3’ truncated fbfgene<br />
with 5’ regions of variable length. The truncated<br />
genes were fused to the promotorless hybrid indicator<br />
gene ∆trp-lacZ and the neo gene for selection.<br />
β-Galactosidase activity increased 8 or 14 hours<br />
after the beginning of starvation in the merodiploid<br />
strains but not during indole induced sporulation. This<br />
unequivocally proves the four fbf-genes to be involved<br />
in the morphogenic process of fruiting. RT-PCR analyses<br />
of fbf-gene transcription revealed these genes to<br />
be induced during starvation. Low levels of fbf-gene<br />
transcript are found in vegetative cell and in the case<br />
of fbfC or fbfD during indole induced sporulation.<br />
Analysis of the protein patterns of the wild-type and<br />
the mutant strains by 2D electrophoresis is in progress.<br />
Alternative sigma factors<br />
Barbara Silakowski, Susanne Müller and Chi-Hyuk<br />
Chang<br />
The genes of two alternative sigma factors, sigB and<br />
sigC have been cloned. These sigma factors harbour<br />
two domains that were shown for σ 32 of E. coli to<br />
be necessary for DnaK binding and thus for its proteolytic<br />
clearance. Merodiploid strains containing the<br />
wild type gene and the corresponding 3’ truncated<br />
gene fused to an indicator gene were analysed for the<br />
expression of the sigma factor genes during development<br />
or heat shock. sigB was shown to be expressed<br />
early during indole-induced sporulation and fruiting<br />
body formation as well during heat shock. These<br />
results agree with those of the RT-PCR analysis of<br />
sigB transcription. Inactivation of either sigB or sigC<br />
by insertional mutagenesis did not impair fruiting<br />
body formation, indole-induced sporulation or the<br />
heat shock response. No changes in either the spores’<br />
ultrastructure (H. Lünsdorf, GBF, Braunschweig) or<br />
in spore germination have been detected.<br />
A gene cluster of S. aurantiaca DW4/3-1 for<br />
Myxothiazol biosynthesis<br />
H. Ehret and B. Silakowski in collaboration with H.<br />
Reichenbach, GBF, Braunschweig<br />
Sequence analyses downstream of the developmental<br />
fbfB gene resulted in the detection of the mta (myxothiazol)<br />
gene cluster. The first ORF, mtaA, (formerly designated<br />
hesA), encodes a phosphopantetheinyl transferase.<br />
P-pant transferases activate polyketide synthases<br />
(PKS) by the transfer of the P-pant moiety from<br />
coenzyme A to a conserved serine residue of the PKS.<br />
Downstream of mtaA a second ORF, mtaB, (formerly<br />
pksA) was found, of which 7 kbp were sequenced. It<br />
encodes a PKS. Inactivation of mtaA by insertional<br />
mutagenesis or deletion of part of the gene or insertional<br />
inactivation of mtaB gene impairs myxothiazol<br />
synthesis. In addition mutants defective in mtaA fail to<br />
form a sofar unknown metabolite. This suggests MtaA<br />
to be able to activate at least two different polyketide<br />
synthases.<br />
For studying the expression of the mtaB gene a merodiploid<br />
strain BS64 was constructed that harboured<br />
a functional wildtype and a 3‘ truncated mtaB gene<br />
to which an indicator gene was fused. Measurement<br />
of indicator gene expression showed, mtaB to be<br />
expressed un<strong>der</strong> all conditions tested, such as vegetative<br />
growth, fruiting, indole induced sporulation and<br />
heat shock. The project was stopped at this stage at the<br />
<strong>ZMBH</strong>. Further investigations of the gene cluster are<br />
performed at the GBF in Braunschweig.<br />
CspA, a prominent cold-shock(-like) protein<br />
I. Stamm and W. Plaga<br />
Several related proteins of about 7 kDa constitute a<br />
prominent fraction of the S. aurantiaca cell extract.<br />
One of the genes encoding such a protein was cloned<br />
and named cspA. The delineated protein sequence of<br />
68 amino acid residues displays a high sequence identity<br />
with bacterial cold-shock(-like) proteins. A RNA<br />
chaperon-function was proposed for these proteins<br />
in E. coli. Using a cspA::(trp-lacZ) fusion gene that<br />
was introduced into Stigmatella by electroporation the<br />
transcription was analyzed during development and<br />
at lowered temperature. These experiments indicated<br />
cspA to be constitutively transcribed at a high level.<br />
The cspA promoter was used to express the gene for<br />
the green fluorescent protein (GFP). GFP fluorescence<br />
was found to be detectable in whole fruiting bodies<br />
as well as in single cells. The GFP-labelled cells are<br />
easily distinguished from wild type cells using a fluorescence-activated<br />
cell sorter (FACS). Fruiting body<br />
formation is not impaired in gfp expressing wild type<br />
strains. Thus the gfp gene un<strong>der</strong> the control of the<br />
cspA promoter seems to be suitable to label Stigmatella<br />
strains. With this kind of labelling it should be<br />
possible to analyze the fate of mutant strain cells<br />
in phenotypic complementation experiments during<br />
fruiting body formation; in such experiments fruiting<br />
bodies are formed by the combined action of two<br />
mutant strains which are unable to develop properly<br />
on their own.<br />
111
Fluorescence based analysis of gene expression:<br />
Identification of pheromone target<br />
genes<br />
The pheromone stigmolone (2,5,8-trimethyl-8-hydroxy<br />
-nonan-4-one) is instrumental in early steps of fruiting<br />
body formation. To identify stigmolone-responsive<br />
genes a promoter trap vector (pTRAP1) was constructed<br />
which allows the creation of random promoter<br />
fusions to gfp in S. aurantiaca. With the aid<br />
of a flow cytometer a selection strategy exploiting the<br />
differential fluorescence induction (DFI) of these promoter<br />
fusions by stigmolone is feasible. First analyses<br />
of the random insertion mutants by flow cytometry<br />
have shown, that about 2.5% of the mutants express<br />
the gfp during vegetative growth. Screening for genes<br />
affected by the addition of stigmolone is in progress.<br />
HspA (SP21): Transcriptional regulation and<br />
biological function<br />
Hui Shen<br />
HspA (formerly SP21) of S. aurantiaca is synthesised<br />
during development or un<strong>der</strong> stress such as heat shock,<br />
oxygen limitation or indole treatment. Sequence alignment<br />
revealed HspA to belong to the α-crystallin<br />
family of the low molecular weight heat shock proteins.<br />
HspA is located mainly at the cell periphery in<br />
heat shocked cells and in fruiting body <strong>der</strong>ived myxospores<br />
and either at the cell periphery or within the<br />
cytoplasma of indole treated cells as shown by immunoelectron<br />
microscopy (H. Lünsdorf, GBF, Braun–<br />
schweig).<br />
Two transcripts of the hspA gene were detected after<br />
heat shock and only one after indole treatment. A<br />
unique transcription initiation site of the monocistronic<br />
hspA gene was detected by primer extension<br />
either after heat shock or indole treatment. Analysis<br />
112<br />
of the promoter proved various upstream regions to<br />
be required for maximum expression of hspA un<strong>der</strong><br />
stress conditions. For maximum hspA transcription<br />
225 bp and 587 bp upstream of the ATG start codon<br />
are required in the case of heat shock or indole treatment,<br />
respectively.<br />
To delimit the regulatory elements involved in hspA<br />
transcription that depends on heat shock, deletion<br />
/ insertion mutagenesis as well as gel shift assays<br />
were performed. Three regulatory promoter regions<br />
involved in heat shock response were defined. The first<br />
region spans from bp -56 to bp -85 upstream of the<br />
hspA ORF and harbours the RNA polymerase binding<br />
sites. Deletion of this region completely blocks hspA<br />
transcription. The second domain ranges from bp -141<br />
to bp -223 upstream of the hspA ORF and carries putative<br />
regulator-binding sites. Heat shock and phosphorylation<br />
enhance binding of the regulator(s) to the hspA<br />
promoter. Deletion of this region reduces hspA transcription<br />
by more than half. Deleting a sequence ranging<br />
from bp -86 to bp -140 upstream of the hspA ORF<br />
abolishes hspA transcription suggesting a cis-acting<br />
element to exist just upstream of the -35, -10 regions<br />
of the hspA promoter. These results suggest the transcription<br />
of hspA to be mainly positively regulated<br />
un<strong>der</strong> heat shock conditions.<br />
A His-tagged fusion protein of Hspa (HspA His ) was<br />
produced in E. coli. This polypeptide tends to assemble<br />
into a large complex that consists of 26 subunits<br />
with a molecular mass of 560 kDa as judged by size<br />
exclusion chromatography. This oligomer of HspA His<br />
interacts with unfolded cytrate synthetase (CS) and<br />
prevents the enzyme‘s precipitation. The unfolded<br />
B-chain of insulin is not protected from precipitation.<br />
A stable complex is formed between HspA His and<br />
unfolded CS because the unfolded enzyme does not<br />
dissociate from the complex. Though thermotoler-<br />
ance and differentiation of S. aurantiaca cells are not<br />
affected in the absence of this protein, one may suggest<br />
HspA to play a protective role in vivo.<br />
II. Molecular Biology of the Infection Process<br />
by the Entomopathogenic Fungus Beauveria<br />
bassiana<br />
E. Duperchy, H. Wan, G. Mitina and A. Leclerque in<br />
cooperation with P. Zink and G. Zimmermann (Bioassays),<br />
Biologische Bundesanstalt f. Land- und Forstwirtschaft,<br />
Darmstadt<br />
The organism<br />
The filamentous fungus Beauveria bassiana (Balsamo)<br />
Vuillemin belongs to a class of insect pathogenic<br />
hyphomycetes (fungi imperfecti). The different<br />
Beauveria strains are highly adapted to particular host<br />
insects. A broad range of B. bassiana species is found<br />
world wide un<strong>der</strong> very different climatic conditions.<br />
The different Beauveria strains cover a rather wide<br />
spectrum of insect hosts that are of medical or agricultural<br />
significance. Hosts of medical importance<br />
include vectors for agents of tropical infectious diseases<br />
such as the tsetse fly Glossina morsitans morsitans,<br />
the sand fly Phlebotomus that transmits Leishmania,<br />
and bugs of the genera Triatoma and Rhodnius,<br />
the vectors of Chagas’ disease. Hosts of agricultural<br />
significance include the Colorado potato beetle<br />
Leptinotarsa decemlineata, the codling moth Carpocapsa<br />
pomonella and several genera of termites.<br />
In the absence of the specific insect host, the facultative<br />
insect pathogen passes through an asexual vegetative<br />
life cycle that includes germination, filamentous<br />
growth, and the formation of sympoduloconidia from<br />
poorly differentiated conidiophores. In the presence of<br />
its host insect, Beauveria switches to the pathogenic<br />
life cycle. The conidiospores germinate on the surface<br />
of the cuticle; the newly generated hyphae penetrate<br />
the insect’s integument and liberate single cells, socalled<br />
blastospores, or hyphal bodies, when reaching<br />
the hemocoel. The hemolymph distributes the blastospores<br />
through the whole body cavity, that thus form<br />
points of origin of new hyphae that invade all host tissues.<br />
When the nutrients of the carcass are used up,<br />
a thick layer of aerial hyphae is formed on the surface<br />
of the insect’s cadaver, of which conidiospores<br />
are released.<br />
During the infectious process fungal structures are<br />
subject to several defense response mechanisms of the<br />
host-insect. These include humoral as well as hematocytic<br />
encapsulation reactions, hemolymph clotting<br />
reactions with the aim to hin<strong>der</strong> blastospore propagation,<br />
and the action of defensin-like small antifungal<br />
peptides.<br />
To overcome the host’s defense mechanisms and physical<br />
barriers it is suggested the parasite to have two<br />
classes of virulence factors. One class comprises the<br />
more general virulence factors that are independent<br />
of the host insect. They include hydrophobin like proteins<br />
that mediate spore adhesion to cuticular structures,<br />
lytic enzymes such as proteases or chitinases for<br />
cuticule penetration and hemocoel invasion and diffusible<br />
cyclopeptide- or depsipeptide-toxins that suppress<br />
the defense reactions of the host. The second<br />
class comprises factors that are needed for the switch<br />
from saprophytic growth to the growth on a specific<br />
host-insect. These factors include a specific receptor-<br />
and signal-transduction-system that allows the fungus<br />
to differentiate between its correct host and other<br />
insects.<br />
113
Characterisation of genes involved in B. bassiana<br />
virulence<br />
For the identification of virulence factors of a B. bassiana<br />
strain that is highly adapted to the Colorado potato<br />
beetle, two complementary strategies are employed.<br />
Firstly, in a REMI (“restriction enzyme mediated integration”)<br />
transformation approach mutants are generated.<br />
Mutants that result from a monolocal recombination<br />
event are tested for their ability to grow on their<br />
host-insect. In the case of a significant reduction of<br />
their virulence the disrupted genomic region will be<br />
cloned and sequenced. Secondly, a mRNA differential<br />
display analysis is performed to identify fungal genes<br />
that are transcriptionally up regulated by the interaction<br />
with the host using cultures grown in the absence<br />
or presence of host structures These genes are cloned<br />
from a wild type genomic DNA library. The significance<br />
of the thus found potential virulence factor<br />
genes for pathogenesis are tested by site-specific inactivation<br />
of the single cloned genes or of groups of<br />
functionally related genes and the determination of<br />
the pathogenicity phenotype of the corresponding null<br />
mutants.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (Graduiertenkolleg “Biotechnologie”, Gradu–<br />
iertenkolleg “Kontrolle <strong>der</strong> Genexpression in pathogenen<br />
Mikroorganismen” and Projekt-’Sachbeihilfen’)<br />
and from the DAAD.<br />
PUBLICATIONS<br />
Silakowski, B., Ehret, H. and Schairer, H.U. (1998).<br />
FbfB, a gene encoding a putative galactose oxidase, is<br />
114<br />
involved in S. aurantiaca fruiting body formation. J.<br />
Bacteriol. 180, 1241-1247.<br />
Zhang, J., Schairer, H.U., Schnetter, W., Lereclus, D.<br />
and Agaisse, H. (1998). Bacillus popilliae cry18Aa<br />
operon is transcribed by σ E and σ K forms of RNA<br />
polymerase from a single initiation site. Nucleic Acids<br />
Res. 26, 1288-1293.<br />
Börcsök, I., Schairer, H.U., Sommer, U., Wakley, G.K.,<br />
Schnei<strong>der</strong>, U., Geiger, F., Niethard, F.U., Ziegler, R.,<br />
and Kasperk, C.H. (1998). Glucocorticoids regulate<br />
the expression of the human osteoblastic Endothelin A<br />
receptor gene. J. Exp. Med. 188, 1563-1573.<br />
Plaga, W., Stamm, I., and Schairer, H.U. (1998). Intercellular<br />
signaling in Stigmatella aurantiaca: Purification<br />
and characterization of stigmolone, a myxobacterial<br />
pheromone. Proc. Natl. Acad. Sci. USA 95,<br />
11263-11267.<br />
Hull, W.E., Berkessel, A., and Plaga, W. (1998). Structure<br />
elucidation and chemical synthesis of stigmolone,<br />
a novel type of prokaryotic pheromone. Proc. Natl.<br />
Acad. Sci. USA 95, 11268-11273.<br />
Stamm, I., Leclerque, A., and Plaga, W. (1999). Purification<br />
of cold-shock-like proteins from Stigmatella<br />
aurantiaca – molecular cloning and characterization<br />
of the cspA gene. Arch. Microbiol. 172, 175-181.<br />
Plaga, W., and Schairer, H.U. (1999). Intercellular signalling<br />
in Stigmatella aurantiaca. Curr. Opin. Microbiol.<br />
2, 593-597.<br />
Plaga, W., Vielhaber, G., Wallach, J., and Knappe, J.<br />
(<strong>2000</strong>). Modification of Cys-418 of pyruvate formate-<br />
lyase by methacrylic acid, based on its radical mechanism,<br />
FEBS Lett. 466, 45-48.<br />
Silakowski, B., Schairer, H.U., Ehret, H., Kunze, B.,<br />
Weinig, S., Nordsiek, G., Brandt, P., Blöcker, H.,<br />
Höfle, G., Beyer, S. and Müller, R. (1999). New lessons<br />
for combinatorial biosynthesis from myxobacteria:<br />
the myxothiazol biosynthetic gene cluster of<br />
Stigmatella aurantiaca DW4/3-1. J. Biol. Chem. 274,<br />
37391-37399.<br />
White, D. and H.U. Schairer (1999). Development of<br />
Stigmatella. In Procaryotic Development. Y.V. Brun<br />
and L. J. Shimkets edts. American Society for Microbiology,<br />
Washington, DC pp 285-294.<br />
THESES<br />
Dissertation<br />
Shen, Hui (1999). Transcriptional regulation of hspA<br />
gene in Stigmatella aurantiaca and function analysis<br />
of HspA protein. Universität Heidelberg.<br />
STRUCTURE OF THE GROUP<br />
E-mail: hus@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Schairer, Hans Ulrich, Prof. Dr.<br />
Postdoctoral<br />
fellows Plaga, Wulf, Dr.<br />
Silakowski, Barbara, Dr.*<br />
Visiting scientist Mitina, Galina, Dr.*<br />
Ph.D. students Chang, Chi-Hyuk, Dipl. Biol.*<br />
Duperchy, Esther, Dipl. Biol.*<br />
Hofmann, Diana, Dipl. Biol.*<br />
Leclerque, Andreas, Dipl. Chem.*<br />
Müller, Susanne, Dipl. Biol.<br />
Shen, Hui, Dipl. Biol.<br />
Wan, Hong, Dipl. Biol.*<br />
Techn. assistant Stamm, Irmela<br />
* part of the time reported<br />
115
Heinz Schaller<br />
Regulation of Hepatitis B Virus Replication<br />
Hepatitis B viruses (also hepadnaviruses, HBVs) are<br />
small, enveloped DNA viruses which cause acute and<br />
chronic liver infections in mammals and birds. Their<br />
prototype, the human HBV, is the causative agent of<br />
a world-wide major public health problem with about<br />
5 % of the world population being chronic HBV carriers<br />
and at high risk of developing liver cirrhosis and<br />
hepatocellular carcinoma. As ‘pararetroviruses’, these<br />
viruses are related to the retroviruses by genome organization<br />
and replication strategy, but differ in major<br />
features, e.g. by containing a DNA genome in the<br />
extra-cellular state, or by producing transcripts from<br />
a non-integrated, circular DNA template. The hepadnaviruses<br />
are characterized by a tissue tropism and<br />
a very narrow host range restricting them to their<br />
respective natural hosts and close relatives. These features<br />
have been an obstacle to the establishment of an<br />
HBV cell culture infection system, which is urgently<br />
needed for the development of targeted therapy.<br />
Despite these experimental difficulties with the HBV<br />
prototype, the hepadnavirus infection cycle has been<br />
worked out in consi<strong>der</strong>able detail from molecular<br />
analysis of HBV replication in cells transfected with<br />
cloned viral DNA, as well as from infection studies<br />
in the duck Hepatitis B virus (DHBV) animal system.<br />
The resulting replication model (Fig. 1) is well estab-<br />
Figure 1. The HBV replication cycle. Individual steps are indicated: (1,2) attachment to, and endocytosis into the host cell;<br />
(3,4) cytosolic release of the capsid and transport to the nucleus; (5) conversion of the viral genome into ccc-DNA, the<br />
template for transcription; (6) synthesis of genomic RNA, its maturation and transport to the cytoplasm, and (7) its translation<br />
into the core and polymerase gene products, followed by (8), coassembly into RNA-containing nucleocapsids, ((8‘)<br />
cyto-nucleoplasmic shuttling of core protein subunits, postulated to participate in genomic RNA maturation); (9) reverse<br />
transcription of the RNA genome into DNA, and (10) nucleocapsid maturation and export from the cell as enveloped virion.<br />
Alternatively, nucleocapsids import the mature DNA genome into the nucleus for replenishment of the ccc-DNA pool (11).<br />
For simplicity, synthesis and assembly of the envelope proteins from subgenomic RNA are omitted.<br />
116<br />
lished with respect to the basic mechanisms leading to<br />
the production of progeny virions (steps 7-10), but still<br />
uncertain as to the mechanisms and cellular components<br />
involved in determining receptor-mediated virus<br />
uptake (steps 2-4), and also as to those that control the<br />
establishment and persistence of productive infection<br />
(primarily steps 4, 6, and 11). These latter processes<br />
are difficult to study un<strong>der</strong> controlled conditions<br />
since no infectable cell lines exist even in case of<br />
the available animal models. Nevertheless, we have<br />
been increasingly focussing our research onto these<br />
research areas, as their better un<strong>der</strong>standing appears<br />
to be crucial for a rational approach towards our longterm<br />
goal, which is to establish an infection system for<br />
studying (and to interfere with) all steps of HBV replication<br />
cycle of the human virus in cultured cells, and<br />
in a small test animal.<br />
I. Parameters influencing the establishment<br />
and maintenance of hepadnavirus replication<br />
in primary hepatocyte cultures<br />
U. Klöcker, B. Zachmann-Brand, B. Glass, C.<br />
Kuhn, K. Rothmann, U. Protzer<br />
Hepatitis B viruses use a well-balanced replication<br />
strategy and an intimate cross talk with the host to<br />
establish productive, but non-cytotoxic, long-term persistent<br />
infections. As part of this strategy, virus replication<br />
and gene expression vary greatly in response<br />
to environmental changes of the state of the cell. For<br />
example, transfer of preinfected duck hepatocytes out<br />
of the tissue structure into cell culture results in an initial<br />
reduction of virus production followed by upregulation<br />
(Fig. 2). Furthermore, extracellular stimuli such<br />
as cytokines, the peptide hormone glucagon, or endotoxin<br />
from gram negative bacteria inhibit the establishment<br />
of DHBV replication in vitro. We have now<br />
shown that endotoxin acts indirectly by activating liver<br />
Figure 2. Changes in virus production from DHBV-infected<br />
primary duck hepatocytes in response to environmental<br />
changes. After plating, preformed virus is released, followed<br />
initially by decreased rates of production. From<br />
day 4 on, virus production increases, reaching constant<br />
levels around day 8. This process is inhibited by endotoxin-induced<br />
cytokines produced from liver macrophages<br />
(Klöcker et al., <strong>2000</strong>).<br />
macrophages to release cytokines which act at inhibitory<br />
an early step of DHBV replication (Fig. 2).<br />
Another example for cross talk with the host is our<br />
finding that the large envelope protein, although a<br />
structural protein, is varibly phosphorylated in cytosolic<br />
preS domains by ERK-type MAP kinases in<br />
response to to the state of the cell. In turn, changes in<br />
preS-phosphorylation correlated closely with the ability<br />
of DHBV L to activate gene expression in trans,<br />
in vitro and in vivo. Furthermore, a pathogenic phenotype<br />
with severe growth retardation and pathologic<br />
liver histology was observed in ducklings infected<br />
with a variant mimicking constitutive L phosphorylation.<br />
The above observations from experimental DHBV<br />
infection are complemented by data obtained with<br />
HBV-transgenic mice, which produce HBV from a<br />
chromosomally integrated HBV genome in titers comparable<br />
to chronically infected humans. In this system,<br />
117
the HBV genome is expressed only in a particular<br />
subset of hepatocytes, again demonstrating the importance<br />
of liver architecture and of hepatocyte microenvironment.<br />
Analysis of virus replication in HBVtransgenic<br />
mice lacking the large envelope protein do<br />
not support a prominent role of the L protein for regulating<br />
the levels of intracellular replication intermediates<br />
in HBV, in contrast to evidence for such a mechanism<br />
for the DHBV system.<br />
II. Hepadnavirus replication after transfer of<br />
hepadnaviral genomes mediated by adenoviral<br />
vectors<br />
M. Sprinzl, H. Oberwinkler, B. Zachmann-Brand,<br />
J. Dumortier, U. Protzer<br />
To bypass the species barrier that precludes animal<br />
studies with the human HBV, we have established<br />
recombinant adenoviruses that transfer HBV and<br />
DHBV genomes into heterologous hepatocytes, or<br />
into cell lines which are not infectable but capable<br />
of supporting virus replication. In contrast to transfection,<br />
this approach allows to introduce a defined<br />
number of hepadnavirus genomes, thereby simulating<br />
natural initiation of hepadnavirus replication from<br />
extrachromosomal DNA templates; it should enable<br />
us to study in more detail in vitro and in vivo the influence<br />
of the state of the host hepatocyte, and the use of<br />
immune mediators on hepadnavirus replication. Thus,<br />
we have started to use adenoviral genome transfer into<br />
cultured mouse hepatocytes to test for the influence of<br />
defined immune mediators (from the mouse) on hepatitis<br />
B virus replication in vitro. Furthermore, transduction<br />
of hepadnavirus genomes was shown to efficiently<br />
establish HBV and DHBV replication in the<br />
mouse liver, yielding serum titers comparable to those<br />
from HBV-transgenic animals. In these mice, viral<br />
118<br />
gene expression and replication was maintained for<br />
up to three months, provided that an (adenovirusdirected)<br />
immune response was suppressed.<br />
III. A Golgi-resident protein participates as<br />
an uptake receptor in duck hepatitis B<br />
virus infection<br />
K. Breiner, B. Glass, S. Urban<br />
Major efforts through two decades have produced a<br />
growing list of HBV receptor candidates, mostly only<br />
defined as virus binding proteins, but have failed to<br />
characterize any of these functionally. More progress<br />
has been made in the DHBV animal model which<br />
allows infection experiments with primary hepatocytes.<br />
Including this essential complement for analysis,<br />
we have now identified and characterized in detail<br />
a bona fide DHBV receptor molecule, which plays<br />
a crucial role in a conceptually new mechanism of<br />
viral entry. This protein, a cellular carboxypeptidase<br />
(CPD, historically termed gp180), had been initially<br />
only characterized as a virus binding protein - and met<br />
with scepticism since it lacked the expected tissue tropism,<br />
species specificity, and since is predominantly<br />
concentrated in the trans-Golgi network and not at the<br />
plasma membrane where conceptually viruses must<br />
interact with host cell receptor(s). Our conclusion that<br />
gp180 is nevertheless a functional DHBV receptor<br />
is based on a comprehensive series of experimental<br />
observations: (i) gp180 is the only duck protein that<br />
binds to recombinant DHBV preS, a ligand on the<br />
virus surface previously shown to be essential for<br />
infection, (ii) a preS subdomain, functionally defined<br />
in infection competition experiments, coincides with<br />
the domain determining physical gp180 binding, (iii)<br />
anti-gp180 antibodies, as well as recombinant gp180/<br />
CPD, efficiently block DHBV infection of cultured<br />
duck hepatocytes, (iv) expression of gp180 in a<br />
human hepatoma cell line mediates cellular attachment<br />
and subsequent internalization of fluorescently<br />
labeled DHBV particles into vesicular structures;<br />
gp180 expression does, however, not ren<strong>der</strong> these<br />
heterologous cells permissive for productive DHBV<br />
infection. Furthermore, gp180/CPD is down-regulated<br />
in DHBV-infected cells through intracellular interaction<br />
with the preS ligand, a finding matching our<br />
observation that DHBV infection reduces subsequent<br />
superinfection about 20-fold. Taken together, these<br />
and further data support the model that gp180/CPD<br />
acts as the uptake receptor in a multistep process<br />
(Fig.3). Although predominantly concentrated intracellularly,<br />
the very limited cell surface exposition of<br />
gp180/CPD (varied experimentally by gp180-transducing<br />
adenoviruses) was found to be sufficient to<br />
mediate virus particle uptake by coendocytosis, while<br />
fusion at an endosomal membrane is predicted to<br />
require an (yet unknown) species-specific fusion coreceptor.<br />
Thus, gp180/CPD represents a first example<br />
for a receptor that is virtually absent from the cell surface,<br />
but nevertheless recognized and utilized for virus<br />
Figure 3. Model of cellular gp180 traffic and DHBV entry.<br />
DHBV-complexed gp180 is arrested in the endosome to<br />
allow interaction with a second receptor postulated to trigger<br />
membrane fusion (Breiner et al., <strong>2000</strong>).<br />
internalization leading to productive infection. These<br />
new insights into hepadnaviral entry from the DHBV<br />
model are now being used to re-design the strategies<br />
in our search for the receptor of the medically relevant<br />
human HBV.<br />
IV. Signals for bidirectional cyto-nucleoplasmic<br />
transport in the DHBV capsid protein<br />
H. Mabit, K. Breiner, A. Knaust, B. Zachmann-<br />
Brand<br />
Like other viruses that replicate in the nucleus, the<br />
hepadnaviruses rely on the the cellular cyto-nucleoplasmic<br />
transport machinery to bring the infecting<br />
nucleocapsid to the nucleus. To get insight into the<br />
mechanism operating and the signals used, we have<br />
started to study this process in the DHBV system. By<br />
analyzing the subcellular localization of various segments<br />
of the DHBV core protein (DHBc) fused to<br />
the green fluorescent protein, and by further mutational<br />
analysis, we mapped a single nuclear localization<br />
signal (NLS) in the DHBc sequence. Mutational<br />
inactivation of this sequence prevented DHBc nuclear<br />
targeting (Fig. 4), and in the context of the complete<br />
virus genome, it blocked virus production from<br />
transfected cells. The mutant genome was still capable<br />
of directing the early steps of DHBV infection in<br />
duck hepatocytes, however, compared to wild type,<br />
with delayed gene expression and without infecting<br />
neighbouring cells, both results supporting the interpretation<br />
that the NLS is essential for intracellular<br />
genome amplification and virus production.<br />
Finally, evidence from heterokaryon experiments suggests<br />
that the DHBc sequence contains, in addition to<br />
its NLS, also a nuclear export signal. We propose<br />
that this NES, or a bidirectional shutling signal, counterbalances<br />
NLS function in the productive state of<br />
119
Figure 4. Loss of nuclear import of DHBV core protein<br />
after mutational changes in its nuclear import signal. (A)<br />
Schematic representation of the GPF-fusion used and the<br />
mutation introduced (215RRRKVK220 → RGGEVK). (B)<br />
Cellular distribution of wild type (RRK; left panel) and<br />
mutant (GGE; right panel) in transfected HuH7 cells. GFP<br />
fluorescence in live cells was analyzed using a confocal<br />
microscope, at day 1 post transfection.<br />
the infected cell, thereby preventing abortive nuclear<br />
accumulation of capsids as observed in chronic HBV<br />
patients and in HBV transgenic mice. On the other<br />
hand, we have observed DHBc accumulation in distinct<br />
nuclear dots (particularly early in infection).<br />
These dots also accumulate pregenomic DHBV RNA,<br />
suggesting a role in RNA maturation and nuclear<br />
export.<br />
V. Envelope-independent membrane-binding<br />
preceeds budding and secretion of mature<br />
hepadnaviral nucleocapsids<br />
H. Mabit<br />
Hepadnaviruses are DNA viruses, but as pararetroviruses,<br />
their morphogenesis initiates with the encapsi-<br />
120<br />
dation of a RNA pregenome and these viruses have<br />
therefore evolved mechanisms to exclude immature<br />
nucleocapsids from participating in budding and secretion.<br />
Using the duck virus model and a flotation assay,<br />
we provide evidence that binding of cytoplasmic core<br />
particles to their target membrane is a distinct step<br />
in morphogenesis, discriminating different populations<br />
of intracellular capsids. The membrane-associated<br />
subpopulation contained largely mature, double<br />
stranded DNA genomes and was devoid of detectable<br />
core protein phosphorylation, both features characteristic<br />
for secreted virions. Against expectation, however,<br />
the selective membrane attachment observed did<br />
not require the presence of the large DHBV envelope<br />
protein, whose interaction has been implicated to be<br />
crucial for selective nucleocapsid-membrane interaction.<br />
Furthermore, removal of surface-exposed phosphate<br />
residues from non-floating capsids, did by itself<br />
not suffice to confer membrane affinity. Collectively,<br />
these observations argue for a model in which nucleocapsid<br />
maturation, involving the viral genome, the<br />
phosphorylation state, and the structure of the capsid,<br />
leads to the exposure of a membrane-binding signal as<br />
an initial step crucial for selecting nucleocapsids destined<br />
to be enveloped and secreted.<br />
VI. Recombinant hepadnaviruses as vectors<br />
for hepatocyte-directed gene transfer<br />
U. Protzer, U. Klöcker, A. Frank, in collaboration<br />
with M. Nassal, Med. Uniklinik Freiburg<br />
Being hepatropic and non-cytopathic viruses, the<br />
hepadnaviruses have been envisaged for many years<br />
to be potential candidates for the development of livertargeted<br />
viral vectors. However, due to the compact<br />
organization of the viral genome (3 kb with overlapping<br />
genes and numerous cis-acting sequence ele-<br />
ments), realization of this potential proved to be rather<br />
difficult. We have now demonstrated that, by appropriate<br />
design (i.e. replacement of the S-gene), recombinant<br />
replication-defective HBVs and DHBVs can be<br />
generated which specifically infect hepatocytes, and<br />
transduce and express foreign genes un<strong>der</strong> control of<br />
an endogenous viral promoter. Exclusive expression<br />
of the transgenes in hepatocytes, but not in non-parenchymal<br />
liver cells, confirmed the predicted tissue- and<br />
cell-type specificity.<br />
Being produced at rather high titers (ca.10 8 /ml),<br />
recombinant hepadnaviruses are useful tools for experimental<br />
gene transfer in hepatocyte cultures and for<br />
the study of hepadnaviral infection and its therapeutic<br />
treatment. For example, genetically marked DHBVs<br />
were used to demonstrate superinfection of hepatocytes<br />
with an established hepadnaviral infection. Furthermore,<br />
expression of a type I interferon, a secreted<br />
protein of therapeutic use, from such a vector interfered<br />
with the replication of the resident virus (Fig.5).<br />
These data introduce the use of local cytokine produc-<br />
Figure 5. Therapeutic effect of a recombinant DHBV transducing<br />
an interferon gene. Preinfected duck hepatocytes<br />
were superinfected at various ratios (moi) with rDHBV-<br />
IFN or with rDHBV-GFP as a negative control. The time<br />
course of progeny virus release is shown.<br />
tion by HBV-mediated gene transfer as a new concept<br />
for the treatment of acquired liver diseases, including<br />
chronic hepatitis B. Finally, by developing genetically<br />
marked hepadnaviruses, provide a simple assay to the<br />
identification of infectable cells, and the opportunity<br />
for gene transfer approaches in searching for the missing<br />
DHBV-coreceptor or other missing co-factors.<br />
VII. A spring-loaded topology of the large<br />
envelope protein of Duck hepatitis B<br />
virus<br />
In collaboration with E. Grgacic, Melbourne<br />
The structure and fusion potential of the duck hepatitis<br />
B virus (DHBV) envelope proteins was examined<br />
by treating viral particles with deforming agents<br />
known to release envelope proteins of viruses from<br />
a metastable to a fusion-active state. Exposure of<br />
DHBV particles to low pH triggered a major structural<br />
change in the large envelope protein (L) resulting in<br />
exposure of a new trypsin cleavage site within its<br />
S domain, but without affecting the same region in<br />
the small surface protein (S) subunits. This conformational<br />
change was associated with increased hydrophobicity<br />
of the particle surface most likely arising<br />
from surface exposure of the hydrophobic first transmembrane<br />
domain. In the hydrophobic conformation,<br />
DHBV particles were able to bind to liposomes and<br />
intact cells, while in their absence these particles<br />
aggregated, resulting in viral inactivation. These results<br />
suggest that some L molecules are in a spring-loaded<br />
metastable state which, when released, expose a previously<br />
hidden hydrophobic domain, a transition potentially<br />
representing the fusion active state of the envelope.<br />
121
VIII. A novel mode of high affinity virus-receptor<br />
interaction<br />
S. Urban, in collaboration with G. Multhaup (<strong>ZMBH</strong>),<br />
and C. Schwarz, U.C. Marx, P. Rösch, Lehrstuhl <strong>für</strong><br />
Biopolymere, Universität Bayreuth.<br />
Entry of duck hepatitis B virus (DHBV) is initiated<br />
by specific interaction of a subdomain of about 85<br />
amino acids of the exterior preS domain of the large<br />
viral envelope protein with the cellular entry receptor<br />
gp180/carboxypeptidase D (CPD). This receptor binding<br />
domain is highly variable in sequence, between<br />
DHBV strains and related avian hepadnaviruses, without<br />
significantly affecting receptor interaction as<br />
shown by either binding-competition or infectioncompetition<br />
experiments. These observations had suggested<br />
that the functionality of the DHBV receptor<br />
binding domain might be predominantly determined<br />
by a particular 3D structure rather than by sequential<br />
amino acid motifs.<br />
In a further biochemically analysis, using recombinant<br />
preS polypeptides from E.coli and soluble, truncated<br />
forms of duck CPD from a baculovirus expression<br />
system, the ligand binding site was mapped to an<br />
enzymatically inactive Carboxypeptidase-like repeat<br />
domain, adjacent to the cellular membrane. Ligand<br />
binding to the receptor was characterized by a 1:1<br />
stoichiometry, and shown to induce significant conformational<br />
changes in the receptor. Kinetic analysis by<br />
surface plasmon resonance spectroscopy determined<br />
the complex dissociation constant KD to 1.5 nM, an<br />
extraordinary high affinity compared to other virus<br />
systems. Including a set of mutant preS polypeptides,<br />
this analysis allowed us also to assess contributions of<br />
different preS-sequence elements to complex formation<br />
and complex stability. A conformational analysis<br />
by 2D-NMR supplemented this approach, revealing<br />
the overall structure of the receptor binding site to be<br />
122<br />
largely unstructured, with a low amount of α-helix.<br />
In combination, these data suggest that receptor binding<br />
proceeds in two steps: Initially, a short α-helical<br />
element near the C-terminus of the receptor-binding<br />
domain initiates formation of a low-affinity complex.<br />
This complex is stabilized sequentially, involving<br />
sixty, essentially non-structured amino acids preceding<br />
the helix. We propose that this mechanism of high<br />
affinity receptor interaction, preserving the potential<br />
of the ligand to adapt structure during binding, has<br />
evolved as an alternative strategy to escape immune<br />
surveillance and the evolutionary pressure inherent to<br />
the compact hepadnaviral genome organization.<br />
External Funding<br />
During the period reported, our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
(Sachbeihilfen, and Graduiertenkolleg „Kontrolle <strong>der</strong><br />
Genexpression in Pathogenen Mikroorganismen“), a<br />
grant from the BMBF (För<strong>der</strong>schwerpunkt „Therapie<br />
mit Molekulargenetischen Methoden“ im Programm<br />
„Gesundheitsforschung <strong>2000</strong>“), from the Boehringer<br />
Ingelheim Foundation, and from the Fonds <strong>der</strong> Chemischen<br />
Industrie.<br />
PUBLICATIONS<br />
Hild, M., Weber, O., and Schaller, H. (1998). Glucagon<br />
Treatment Interferes with an Early Step of Duck<br />
Hepatitis B Virus Infection. J. Virol. 72, 2600-2606.<br />
Urban, S., Breiner, K. M., Fehler, F, Klingmüller, U.,<br />
and Schaller, H. (1998). Avian Hepatitis B Virus Infection<br />
is Initiated by Interaction of a Distinct Pre-S Subdomain<br />
with the Cellular Receptor gp180. J. Virol. 72,<br />
8089-8097.<br />
Breiner, K.M., Urban, S., and Schaller, H. (1998).<br />
Carboxypeptidase D (gp180), a Golgi-Resident Protein,<br />
Functions in the Attachment and Entry of Avian<br />
Hepatitis B Viruses. J. Virol. 72, 8098-8104.<br />
Rothmann, K., Schnölzer, M., Radziwill, G., Hildt, E.,<br />
K. Mölling, and Schaller, H. (1998). Host Cell - Virus<br />
Cross Talk: Phosphorylation of a Hepatitis B Virus<br />
Envelope Protein Mediates Intracellular Signaling. J.<br />
Virol. 72, 10138-10147.<br />
Protzer, U., Nassal, M., Chiang, P-W., Kirschfink, M.,<br />
& Schaller, H. (1999). Interferon gene transfer by a<br />
novel hepatitis B virus vector efficiently suppresses<br />
wild-type virus infection. Proc. Natl. Acad. Sci. USA,<br />
96, 10818-10823.<br />
Urban, S., Kruse, C., and Multhaup, G. (1999). A Soluble<br />
Form of the Avian Hepatitis B Virus Receptor. J.<br />
Biol. Chem. 274, 5707-5715.<br />
Grgacic, E.V.L., Kuhn, C. and Schaller, H. (<strong>2000</strong>).<br />
Hepatitis B Virus Envelope Topology: Membrane<br />
Insertion of a Loop Region and Role of S in L Protein<br />
Translocation. J. Virol. 74, 2455-2458.<br />
Breiner, K.M. & Schaller, H. (<strong>2000</strong>). Cellular Receptor<br />
Traffic is Essential for Productive Duck Hepatitis<br />
B Virus Infection. J. Virol. 74, 2203-2209.<br />
Urban, S., Schwarz, C., Marx, U.C., Zentgraf, H.,<br />
Schaller, H., and Multhaup, G. (<strong>2000</strong>). Receptor recognition<br />
by a hepatitis B virus reveals a novel mode of<br />
high affinity virus-receptor interaction. EMBO J. 19,<br />
21227-1227.<br />
Hegenbarth, S., Gerolami, R., Protzer, U., Trans, P.L.,<br />
Brechot, C., Gerken, G., and Knolle, P.A. (<strong>2000</strong>).<br />
Liver sinusoidal endothelial cells are not permissive<br />
for adenovirus type 5. Hum. Gene Ther. 11, 481-486.<br />
Grgacic, E.V.L., and Schaller, H. (<strong>2000</strong>). A springloaded<br />
topology of the large envelope protein of duck<br />
hepatitis B virus: low pH release results in a transition<br />
to a hydrophobic potentially fusogenic conformation.<br />
J. Virol. 74, 5116-5122.<br />
Klöcker, U., Schultz, U., Schaller, H., and Protzer,<br />
U. (<strong>2000</strong>). Liver macrophages release mediators after<br />
endotoxin stimulation that inhibit an early step in<br />
hepadnavirus replication. J. Virol. 74, 5525-5533.<br />
Protzer, U. and Schaller, H. (<strong>2000</strong>). Immune Escape<br />
by Hepatitis B Viruses. in Virus Genes 21, 27-37.<br />
Breiner, K.M., Urban, S., Glass, B., and Schaller, H.<br />
(<strong>2000</strong>) Envelope Protein-Mediated Down-Regulation<br />
of a Hepatitis B Virus Receptor in Infected Hepatocytes.<br />
J. Virol. (in press)<br />
Mabit, H., and Schaller H. (<strong>2000</strong>) Intracellular Hepadnaviral<br />
Nucleocapsids are Selected for Secretion by<br />
Envelope Protein-independent Membrane Binding. J.<br />
Virol. (in press).<br />
THESES<br />
Breiner, Klaus M. (1998): Carboxypeptidase D<br />
(gp180): Rezeptor, Transzeptor und Signalmolekül <strong>für</strong><br />
Vogelhepatitis B Viren.<br />
Rothmann, Kirsten (1998): Kommunikation zwischen<br />
Virus und Wirtszelle: Die Phosphorylierung des grossen<br />
Hüllproteins des Enten Hepatitis B Virus vermit-<br />
123
telt intrazelluläre Signaltransduktion.<br />
Klöcker, Uta (<strong>2000</strong>): Regulation <strong>der</strong> hepadnaviralen<br />
Replikation: Untersuchung des Effektes antiviraler<br />
Mediatoren mit Hilfe hepadnaviraler Vektoren.<br />
Sprinzl, Martin Franz (<strong>2000</strong>): Der adenovirale Genomtransfer<br />
überwindet die Speziesbarriere <strong>der</strong> Hepatitis-<br />
B-Viren.<br />
Habilitations<br />
Hug, Hubert (1998). Signalübertragung <strong>der</strong> CD95<br />
(Fas/APO-1)-vermittelten Apoptose.<br />
Knolle, Percy (<strong>2000</strong>). Die Leber als immunregulatorisches<br />
Organ: Bedeutung <strong>der</strong> sinusoidalen Endothelzellen<br />
und Kupffer Zellen.<br />
Protzer, Ulrike (<strong>2000</strong>). Virus-Wirt-Interaktion bei <strong>der</strong><br />
Hepatitis B Virus Infektion: vom verbesserten Verständnis<br />
zur Entwicklung neuer Therapien.<br />
Urban, Stephan (<strong>2000</strong>). Identifizierung des aviären<br />
Hepatitis B Virus Rezeptors und Charakterisierung<br />
seiner Interaktion mit dem viralen Hüllprotein.<br />
STRUCTURE OF THE GROUP<br />
E-mail: hshd@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Schaller, Heinz, Prof. Dr.<br />
Research associates/<br />
postdoctoral<br />
fellows Knaust, Andreas, Dr. *<br />
Kuhn, Christa, Dr.<br />
Mabit, Hélène, Dr.*<br />
124<br />
Obert, Sabine, Dr.*<br />
Protzer, Ulrike, Dr.<br />
Sirma, Hüseyin, Dr.*<br />
Urban, Stephan, Dr.<br />
Visiting<br />
scientists Wimmer, Eckard, Prof.*<br />
from SUNY at Stony Brook, USA<br />
(Humboldt Awardee)<br />
Grgacic, Elizabeth, Dr.*<br />
from Macfarlane Burnet Centre,<br />
Fairfield, Australia<br />
Ph.D. students Breiner, Klaus, Dipl.Ing.,<br />
MA chem.<br />
Dumortier, Jêrome*<br />
Klöcker, Uta<br />
Rothmann, Kirsten*<br />
Sprinzl, Martin*<br />
Thake, Sandra*<br />
Techn.<br />
assistants Glass, Bärbel, Dipl. Biol.<br />
Götzmann, Martina*<br />
Oberwinkler, Heike*<br />
Zachmann-Brand, Beate<br />
*) part of the time reported<br />
Project Group Percy Knolle<br />
Regulation of the Immune Response in the<br />
Liver<br />
The liver is a unique organ with regard to the induction<br />
of peripheral immune tolerance. Delivery of antigen<br />
into the liver results in tolerance induction towards this<br />
specific antigen as is seen by the increased survival<br />
of organ transplants following portal venous injection<br />
of donor antigens or following portal venous drainage<br />
of the organ transplant. A number of different hepatic<br />
cell populations and the unique hepatic microenvironment<br />
seem to cooperate to induce immune tolerance.<br />
We are studying the contribution to immune tolerance<br />
of the sinusoidal endothelial cells of the liver (LSEC),<br />
which interact constitutively with leukocytes in the<br />
sinusoidal lumen (see Fig. 1) as well as two other<br />
Figure 1. Schematic drawing of the relative position and<br />
cell sizes in the hepatic sinusoid. Leukocytes are in intimate<br />
contact with LSEC during liver passage and constitutively<br />
adhere to LSEC un<strong>der</strong> physiologic conditions.<br />
hepatic sinusoidal cell populations, the Kupffer cells<br />
and the liver associated lymphocytes. Kupffer cells<br />
and LSEC have the capacity to act as antigen-presenting<br />
cells and activate CD4 + T cells suggesting<br />
immune-surveillance function for the liver. However,<br />
antigen-presentation by LSEC and Kupffer cells is<br />
stringently controlled by physiological constituents<br />
of portal venous blood (e.g. endotoxin) and by both,<br />
autocrine and paracrine action of soluble mediators,<br />
which are induced in resident liver cells by endotoxin.<br />
Activation of naive CD4 + T cells by LSEC outside<br />
lymphoid tissue may constitute one mechanism of<br />
hepatic tolerance induction, because antigen-presention<br />
by LSEC leads to the generation of CD4 + T cells<br />
that express immunosuppressive cytokines and act as<br />
regulatory T cells.<br />
I. Functional inactivation of CD8+ T cells<br />
by cross-presenting sinusoidal endothelial<br />
cells<br />
J. Ohl und A. Limmer<br />
Presentation of endocytosed antigens on MHC-class<br />
I molecules on antigen-presenting cells to cytotoxic<br />
CD8 + T cells (cross-presentation) is important for<br />
immunity to intracellular pathogens and was supposed<br />
to be restricted to dendritic cells and macrophages.<br />
Once in the blood, antigen is taken up efficiently by<br />
LSEC, but not significantly by other cell populations<br />
of the liver (Fig. 2). In vitro analysis of isolated<br />
LSEC reveiled that cross-presentation of endocytosed<br />
antigen to a antigen-specific CD8 + T cell hybridoma<br />
occured in LSEC. Cross-presentation in LSEC compared<br />
to other antigen-presenting cells requires only<br />
125
small numbers of antigen-molecules. Dependence of<br />
cross-presentation on the proteasom and TAP suggests<br />
that LSEC possess an efficient mechanism to deliver<br />
endocytosed antigen from the endosom to the cyto-<br />
Figure 2. Antigen uptake by LSEC in vivo and in vitro. (A) Mice<br />
were intravenously injected with 100 µg of Texas-red labelled<br />
ovalbumin and antigen-uptake into liver cells was examined<br />
by confocal microscopy of perfusion fixed liver slices. Antigen<br />
accumulates in a sinusoidal cell population which can be identified<br />
as LSEC by specific uptake of acetylated LDL. (B) Isolated<br />
LSEC were incubated with 10 µg/ml of Texas-red labelled ovalbumin<br />
and examined for antigen-uptake by confocal microscopy<br />
1 hour later. Ovalbumin accumulates in vesicular structures in<br />
LSEC; note the absence of a diffuse cytoplasmic distribution of<br />
ovalbumin typically observed in dendritic cells.<br />
126<br />
plasm for antigen-degradation and MHC-class I peptide-loading.<br />
Further evidence for the contribution of<br />
LSEC to immune tolerance in the liver comes from<br />
the observation that CD8 + T cells activated by crosspresenting<br />
LSEC loose their capacity to express Interferon<br />
γ and loose their cytotoxic activity towards<br />
specific target cells. Thus, LSEC seem to contribute<br />
to hepatic immune tolerance by inducing regulatory<br />
CD4 + T cells and silencing CD8 + T cell mediated<br />
immune responses. Further experiments concentrate<br />
on the role of LSEC in tolerance induction in vivo using<br />
a newly established method of orthotopic implantation<br />
of LSEC into mice.<br />
II. Sinusoidal endothelial cells are not permissive<br />
for adenovirus type 5 infection<br />
S. Hegenbarth and P. Knolle<br />
Adenoviral gene therapy vectors efficiently transduce<br />
hepatocytes in vivo and are already successfully<br />
employed to correct hepatic metabolic disor<strong>der</strong>s. Little<br />
is known, however, on the susceptibility to adenovirus<br />
infection of non-parenchymal liver cells, such as<br />
LSEC. For instance, LSEC express Factor VIII and<br />
are therefore a relevant gene therapeutic target for correction<br />
of Factor VIII deficiency. We have shown, that<br />
LSEC of different species (mouse, tupaia berlenghi<br />
and human) are not permissive for infection with adenovirus<br />
type 5 in vitro or in vivo. In cocultures of<br />
hepatocytes and LSEC, no infection of LSEC was<br />
observed at virus titers that infected 100% of hepatocytes<br />
(Fig. 3). Employing extremely high virus titers,<br />
that could never be obtained in vivo, only single LSEC<br />
were transduced by adenovirus in vitro (Fig. 3). Macrovascular<br />
endothelial cells, however, were efficiently<br />
infected by adenovirus in vitro (Fig. 3). The apparent<br />
Figure 3. Lack of infection of LSEC by Ad5.CMV-GFP.<br />
(A) Cocultures of murine hepatocytes and LSEC were incubated<br />
for one hour with Ad5.CMV-GFP and examined<br />
for GFP-expression 24 hours later. LSEC were identified<br />
by uptake of DiI-acetylated LDL. No infection of LSEC<br />
by Ad5.CMV-GFP is observed. Hepatocytes are efficiently<br />
infected and express GFP. (B). Isolated LSEC were incubated<br />
with Ad5.CMV-GFP at 5.000 infectious units per cell<br />
for one hour. Infection of single LSEC was observed after<br />
24 hours. LSEC identification by uptake of DiI-ac-LDL. (C)<br />
Macrovascular endothelial cells at passage 3 were incubated<br />
with Ad5.CMV-GFP at a concentration of 5 infectious<br />
units per cell. More than 70% of endothelial cells<br />
express GFP after 24 hours as a marker of productive infection.<br />
lack of LSEC infection by adenovirus may be the<br />
consequence of the absent expression of the integrin<br />
α v β 3 , which is important for adenovirus cell entry.<br />
Two other viral vectors, Herpes simplex virus and lentivirus<br />
based vectors, do not productively infect LSEC<br />
either. We conclude that modified or new vectors are<br />
needed for successful targeting of gene expression to<br />
LSEC.<br />
III. Role of sinusoidal endothelial cells for<br />
DHBV infection of hepatocytes<br />
In cooperation with K. Breiner from the FG of<br />
H. Schaller<br />
Infection of hepatocytes by hepatotropic viruses is<br />
thought to occur via specific hepatocellular receptors.<br />
We studied the contribution of LSEC to infection<br />
of hepatocytes by the avian hepaDNA virus, DHBV.<br />
Although the primary binding and uptake receptor<br />
for DHBV-infection of hepatocytes is known (gp180),<br />
its ubiquitous expression appears to contradict its<br />
involvement in viral hepatocyte targeting. We have<br />
found that blood borne DHBV-particles are endocytosed<br />
and accumulate in LSEC but not in hepatocytes<br />
both, in vivo and in vitro. The preferential uptake of<br />
DHBV-particles by LSEC is not compatible with<br />
passive diffusion of viral particles through endothelial<br />
fenestrae as the mechanism of hepatocyte infection.<br />
Once endocytosed, viral particles colocalize with<br />
gp180 in LSEC suggesting a function of the receptor<br />
not only in hepatocyte infection but also in intracellular<br />
trafficking in LSEC. To comply with low dose infection,<br />
we propose that endocytosed viral particles are<br />
transported with the viral receptor through LSEC to<br />
infect neighbouring hepatocytes.<br />
127
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (Graduiertenkolleg “Kontrolle <strong>der</strong> Genexpression<br />
in pathogenen Mikroorganismen” and Projekt-<br />
’Sachbeihilfen’) and from Bayer AG.<br />
PUBLICATIONS<br />
Knolle, P.A., S. Kremp, T. Hohler, F. Krummenauer,<br />
P. Schirmacher and G. Gerken (1998). Viral and host<br />
factors in the prediction of response to interferon-<br />
alpha therapy in chronic hepatitis C after long-term<br />
follow-up. J. Viral. Hepat. 5 (6): 399-406.<br />
Knolle, P.A., A. Uhrig, S. Hegenbarth, E. Loser, E.<br />
Schmitt, G. Gerken and A. W. Lohse (1998). IL-10<br />
down-regulates T cell activation by antigen-presenting<br />
liver sinusoidal endothelial cells through decreased<br />
antigen uptake via the mannose receptor and lowered<br />
surface expression of accessory molecules. Clin. Exp.<br />
Immunol. 114 (3): 427-33.<br />
Knolle, P.A., A. Uhrig, U. Protzer, M. Trippler, R.<br />
Duchmann, K.H. Meyer zum Büschenfelde and G.<br />
Gerken (1998). Interleukin-10 expression is autoregulated<br />
at the transcriptional level in human and murine<br />
Kupffer cells. Hepatology 27 (1): 93-9.<br />
Knolle, P. A., T. Germann, U. Treichel, A. Uhrig, E.<br />
Schmitt, S. Hegenbarth, A. W. Lohse and G. Gerken<br />
(1999). Endotoxin Down-Regulates T Cell Activation<br />
by Antigen-Presenting Liver Sinusoidal Endothelial<br />
Cells. J. Immunol. 162 (3): 1401-1407.<br />
Knolle, P. A., E. Schmitt, S. Jin, T. Germann, R. Duch-<br />
128<br />
mann, S. Hegenbarth, G. a. Gerken and A. Lohse<br />
(1999). Liver sinusoidal endothelial cells can prime<br />
naive CD4+ T cells in the absence of IL-12 and induce<br />
IL-4 production in primed CD4+ T cells: Implications<br />
for tolerance induction in the liver. Gastroenterology<br />
116 (6): 1428-40.<br />
Hegenbarth, S., R. Gerolami, L. Tran, U. Protzer, C.<br />
Brechot, G. Gerken and P. A. Knolle (<strong>2000</strong>). Liver<br />
sinusoidal endothelial cells are not permissive for<br />
infection with adenovirus type 5. Human Gene Therapy<br />
11, 481-486.<br />
Knolle, P. A. and G. Gerken (<strong>2000</strong>). Local regulation<br />
of the immune response in the liver. Immunological<br />
Reviews 174, 21-34<br />
STRUCTURE OF THE GROUP<br />
E-mail: P.Knolle@<strong>ZMBH</strong>.Uni-Heidelberg.de<br />
Project group lea<strong>der</strong> Knolle, Percy, Priv. Doz. Dr.<br />
Postdoctoral fellow Limmer, Andreas, Dr.<br />
Ph.D. students Ohl, Jutta, Dipl. Biol.<br />
Wingen<strong>der</strong>, Andreas,<br />
Dipl. Biol.*<br />
Techn. assistant Hegenbarth, Silke<br />
*part of the time reported<br />
Blanche Schwappach<br />
Quality Control of Ion Channels and<br />
ABC Proteins<br />
Each cell type possesses an exquisitely regulated set<br />
of plasma membrane proteins. It is essential that these<br />
be expressed in their properly folded and correctly<br />
assembled form in appropriate numbers at the cell surface.<br />
This is particularly true for ion channels since<br />
a very small number of channel proteins can have<br />
dramatic effects on the electrical excitability of a<br />
cell. ATP-binding cassette (ABC) proteins constitute a<br />
large superfamily of transporters present in both pro-<br />
and eukaryotes. They utilize ATP to move substrates<br />
as diverse as metal ions, peptides, and lipids across<br />
membranes of cells and cellular organelles. Channels<br />
and ABC transporters play an important role in human<br />
disease - in inherited disor<strong>der</strong>s where many mutations<br />
in the corresponding genes have been identified,<br />
as drug targets, or a cause of drug resistance. In<br />
some diseases like cystic fibrosis or Liddle’s syndrome<br />
intracellular trafficking of the mutated ABC or<br />
channel protein is known to be abnormal. The same<br />
may be true for some cases of familial hyperinsulinism,<br />
a disor<strong>der</strong> caused by mutations in the genes for<br />
the K ATP channel complex.<br />
Almost all ion channels are multimeric protein complexes<br />
and the respective composition of the complex<br />
can greatly affect its functional properties. How is<br />
assembly and precise stoichiometry of ion channels<br />
controlled? What determines the subcellullar localization<br />
of ion channel and ABC protein subunits at different<br />
stages of assembly? How is the activity of newly<br />
synthesized channel proteins compatible with ionic<br />
homeostasis of the endoplasmic reticulum? How does<br />
the cell regulate the number of channels and transporters<br />
at the cell surface? How are the relevant quality<br />
control and trafficking processes regulated in response<br />
to environmental change?<br />
To address these questions we have started to employ<br />
new methods that expand the traditional set of tools<br />
like site-directed mutagenesis, immunofluorescence,<br />
co-immunoprecipitation, and analysis of glycosylation<br />
patterns. We have developed a very sensitive, quantitative<br />
assay for surface protein in Xenopus oocytes so<br />
that surface trafficking and functional properties can<br />
now be precisely correlated even for low-expressing,<br />
multimeric channel proteins. The assay is the basis of<br />
a novel approach to studying protein-protein interactions<br />
between membrane proteins in a native environment.<br />
Retroviral gene transfer methods in mammalian<br />
cells now make it possible to introduce large cDNA<br />
libraries permanently into diverse cell types. Combination<br />
of this approach with the advanced possibilities<br />
of flow cytometry greatly increases the feasibility of<br />
expression cloning and random screening projects in<br />
mammalian cells. We intend to probe the consensus<br />
and context dependence for known and new trafficking<br />
signals in efficient, unbiased random screens.<br />
Detailed knowledge of these parameters, particularly<br />
for common motifs, will be indispensable to identify<br />
putative trafficking signals in the growing databases.<br />
Background: trafficking of the K ATP channel<br />
complex<br />
To address issues of quality control during ion channel<br />
assembly, we have studied the assembly-dependent<br />
trafficking of ATP-sensitive K+ channels. K ATP channels<br />
couple the metabolic state of the cell to membrane<br />
excitability. They are important in many tissues<br />
and regulate insulin secretion in the pancreas, control<br />
129
vascular tone, protect neurons and muscles from ischemia,<br />
and are responsive to leptin. K ATP channels have<br />
an unusual octameric stoichiometry consisting of four<br />
pore-lining inward rectifier a α subunits (Kir6.1/6.2;<br />
two transmembrane segments) like other K+ channels,<br />
but also contain four regulatory sulphonylureabinding<br />
ß subunits (SUR1/2A/2B; probably seventeen<br />
transmembrane segments) that belong to the ATPbinding<br />
cassette (ABC) family of proteins.<br />
We found that only octameric K ATP channel complexes<br />
were capable of expressing on the cell surface, implying<br />
that quality control mechanisms must exist to prevent<br />
monomers and partial complexes from expressing<br />
on the cell surface. Surprisingly, we found that<br />
the primary quality control mechanism during K ATP<br />
assembly did not involve ER degradation or ER chaperones,<br />
but rather the exposure of an ER retention/<br />
retrieval signal (RKR) present in cytosolic domains<br />
of each subunit. Mutating the retention sequences did<br />
not affect protein levels, but allowed surface expression<br />
of monomers and partially assembled complexes,<br />
including improperly gated channel combinations. We<br />
further showed that the RKR trafficking signal was<br />
functional in a variety of eukaryotic cells including<br />
yeast, mammalian cells, and Xenopus oocytes. Interestingly,<br />
this RKR motif did not require proximity<br />
to the N- or C-terminus like all other known ER<br />
retention/retrieval signals and may, therefore, be more<br />
common. In conclusion, quality control during K ATP<br />
assembly is mediated by a short trafficking signal<br />
whose exposure reflects the assembly state of the<br />
channel. These results provide a clear example of how<br />
a trafficking checkpoint serves as an important quality<br />
control mechanism during the assembly of an ion<br />
channel complex. Furthermore, they identify a new<br />
ER retention/retrieval motif that could explain transient<br />
or permanent ER localization of many proteins.<br />
130<br />
Our current projects are designed to better define this<br />
new class of ER retention/retrieval signals, to extend<br />
the analysis of their role in assembly-dependent trafficking<br />
using K ATP channels as a model system, and<br />
to identify and characterize the molecular machinery<br />
that recognizes this class of signals.<br />
PUBLICATIONS<br />
Schwappach B., Stobrawa S., Hechenberger M., Steinmeyer<br />
K., and Jentsch T.J. (1998). Golgi Localization<br />
and functionally important domains in the NH 2 and<br />
COOH terminus of the yeast CLC putative chloride<br />
channel Gef1p. J. Biol. Chem. 273, 15110–15118.<br />
Zerangue N.*, Schwappach B.*, Jan Y.N., and Jan<br />
L.Y. (1999). A new ER trafficking signal regulates<br />
the subunit stoichiometry of plasma membrane K ATP<br />
channels. Neuron 22, 537-548 (*these authors contributed<br />
equally).<br />
Schwappach, B.*, Zerangue, N.* Jan Y.N., and Jan<br />
L.Y. (<strong>2000</strong>). Molecular basis for K ATP assembly: transmembrane<br />
interactions mediate association of a K +<br />
channel with an ABC transporter. Neuron 26, 155-167<br />
(*these authors contributed equally).<br />
STRUCTURE OF THE GROUP*<br />
E-mail: b.schwappach@zmbh.uni-heidelberg.de<br />
Group lea<strong>der</strong> Schwappach, Blanche, Dr.*<br />
Ph.D. student Yuan, Hebao, M.A.*<br />
Techn. assistant Metz, Jutta*<br />
* since July/August <strong>2000</strong><br />
Dominique Soldati<br />
Cell and Molecular Biology of the Obligate<br />
Intracellular Parasite Toxoplasma<br />
gondii<br />
Apicomplexan parasites are of enormous medical<br />
and veterinary significance, being responsible for a<br />
wide variety of diseases including malaria, toxoplasmosis,<br />
coccidiosis and cryptosporidiosis. Successful<br />
attachment and invasion of the host cells are key to<br />
the survival of these obligate intracellular parasites.<br />
T. gondii, the most ubiquitous of the Apicomplexa,<br />
has developed a remarkable ability to actively penetrate<br />
almost any nucleated cells from virtually all<br />
warm-blooded animals. Most invasive forms of the<br />
Apicomplexa share a common set of apical structures<br />
and exhibit an unusual form of substrate-dependent<br />
gliding motility as an adaptative mechanism to<br />
actively penetrate host cells. In absence of locomotive<br />
organelles such as cilia or flagella, the basic<br />
engine for gliding locomotion is the actin cytoskeleton<br />
and involves myosin(s) to generate the mechanochemical<br />
force along the actin filaments, allowing<br />
parasites to move at rates from 1 to 10 micrometers<br />
per second.<br />
Successive exocytosis of regulated compartments<br />
including rhoptries and micronemes is part of the<br />
invasion process. Micronemal proteins are apparently<br />
used for host-cell recognition, binding, and possibly<br />
motility, while rhoptry and dense granule proteins<br />
contribute to the parasitophorous vacuole formation<br />
and its remodeling into a metabolically active compartment.<br />
Release by micronemes occurs in the earliest<br />
phase of invasion, upon contact with the host cells<br />
and raise in intracellular Ca 2+ .<br />
Molecular motors, adhesins and proteases are among<br />
the distinct classes of genes coding for invasion factors.<br />
These genes are anticipated to be essential for<br />
the survival of these parasites and therefore the ability<br />
to conditionally turn on or off their expression is<br />
prerequisite for their in vivo studies.<br />
I. Myosins: molecular motors in Apicomplexa<br />
parasites<br />
Five myosins of T. gondii and three myosins of P. falciparum<br />
have been identified so far. These unconventional<br />
myosins are the foun<strong>der</strong>s of a novel phylogenetic<br />
and structural class of myosin (XIV), restricted<br />
to the Apicomplexa. Members of this class are small<br />
with molecular weights ranging between 90 and 125<br />
kDa and exhibit unusual structural features.<br />
Myosins have been implicated either genetically, biochemically<br />
or by cytolocalization in a variety of cellular<br />
functions. Three distinct forms of gliding characterize<br />
T. gondii motility: circular gliding, upright<br />
twirling, and helical rotation. Inhibitors of actin filaments<br />
(cytochalasin D) and myosin ATPase (butanedione<br />
monoxime) disrupted all three forms of motility.<br />
When applied on intracellular parasites, these<br />
inhibitors also interfere with proper parasite division.<br />
To determine whether one of these myosins functions<br />
as a motor in actin-based gliding motility and<br />
plays a role in cell replication, we have examined<br />
their stage-specific pattern of expression, subcellular<br />
distribution and biochemical properties.<br />
131
TgMyoA: a candidate motor to power gliding<br />
motility<br />
C. Hettmann, A. Geiter , F, Delbac<br />
T. gondii myosin A (TgMyoA) is constitutively<br />
expressed and localizes beneath the plasma membrane<br />
of the parasite. Thus, TgMyoA is in an<br />
ideal position to transmit mechanical energy into<br />
forward motion propelling parasites into host cells.<br />
We mapped the membrane localization determinant<br />
within the short carboxy-terminal tail of TgMyoA,<br />
and site-directed mutagenesis revealed two essential<br />
arginine residues. The closest homologue in P. falciparum,<br />
PfMyoA is transcribed specifically in the<br />
invasive blood stage form and shows the same subcellular<br />
localization in merozoites. The nature of the<br />
receptor at the plasma membrane remains to be determined.<br />
TgMyoD and TgMyoE are similar to TgMyoA, but<br />
predominantly transcribed in the dormant bradyzoite<br />
stage. We have abrogated the expression of TgMyoD<br />
by double homologous recombination in the tachyzoites<br />
of the virulent RH strain without any apparent<br />
detectable phenotype. We need now to reproduce this<br />
knockout in a persistent strain capable to differentiate<br />
into bradyzoites, in or<strong>der</strong> to analyze the phenotypic<br />
consequences of the absence of TgMyoD in the<br />
appropriate stage.<br />
Biochemical characterization of TgMyoA<br />
A. Herm, in collaboration with M. Geeves (Kent<br />
UK), D. Manstein (MPI, Heidelberg) and E. Meyhöfer<br />
(Hannover)<br />
To power gliding motility, TgMyoA must be able to<br />
track along actin filaments upon ATP hydrolysis at<br />
a speed of 1-3 µm/s. We have determined the biochemical<br />
and physical properties of histidine tagged-<br />
132<br />
TgMyoA purified from culture recombinant parasites.<br />
Transient kinetic analysis of TgMyoA was<br />
obtained by stopped-flow and flash photolysis methods<br />
and revealed strong similarities to typical fast<br />
myosins, like rabbit skeletal myosin. The sliding<br />
velocity of fluorescently labeled actin filaments on<br />
TgMyoA attached to nitrocellulose-coated glass was<br />
5-6 µm/s. A step size of 5 to 6 nm was determined by<br />
single molecule assay monitoring filament displacement<br />
using optical tweezers. TgMyoA shows all the<br />
characteristics of a fast myosin of the class II but<br />
consi<strong>der</strong>ing the significant divergence in the converter<br />
domain and the quasi absence of neck, it is<br />
unclear at the moment how such a motor can generate<br />
a power stroke.<br />
TgMyoB/C: One gene, two tails, and two localizations.<br />
F. Delbac, C. Hettmann, A. Sänger<br />
TgMyoB and TgMyoC are alternative-spliced variants<br />
of the same gene, which produce two myosins<br />
exhibiting different tails. TgMyoB distributes evenly<br />
in the cytosol and is essentially soluble. In contrast,<br />
TgMyoC localizes at the posterior pole of the parasites<br />
describing a ring structure reminiscent of the<br />
termination of the inner membrane complex (IMC).<br />
TgMyoC protein fractionates with membranes and<br />
partitions in the detergent-insoluble fraction. Inhibitors<br />
of myosin ATPase and actin polymerization disrupt<br />
the or<strong>der</strong>ly turnover of mother cell organelles<br />
during daughter cell formation. The circular distribution<br />
of TgMyoC suggests a possible role of this<br />
myosin in the assembly and elongation of the IMCmicrotubule<br />
complex during parasite division. Moreover<br />
the overexpression of TgMyoB slows the rate of<br />
replication and leads to the formation of large resid-<br />
ual bodies, providing an additional indication of the<br />
involvement of TgMyoB/C in cell division.<br />
Figure 1. Immunolocalization of Myosin C (in red) and<br />
MIC6 (in green).<br />
II. Micronemal proteins of several apicomplexan<br />
parasites are sharing common<br />
structural and functional features<br />
The rapid and efficient nature of invasion by Apicomplexa<br />
relies on a sequence of events that are<br />
tightly controlled in time and space. Micronemes are<br />
involved in the trafficking and sequestration of binding<br />
ligands for host cell receptors. These organelles<br />
ensure the appropriate release of ligands in high concentration,<br />
at the tip of the parasite and upon response<br />
to external stimuli which senses contact with the<br />
host cells. To accommodate for the broad host range<br />
specificity of T. gondii, we postulated that adhesion<br />
should involve the recognition of ubiquitous surfaceexposed<br />
host molecules or, alternatively, the presence<br />
of various parasite attachment molecules able<br />
to recognize different host cell receptors. We identified<br />
and characterized a soluble protein MIC4, as<br />
well as a novel family of transmembrane microne-<br />
mal proteins in T. gondii. These proteins are released<br />
at the time of invasion and share domains of homology<br />
with proteins described in other members of the<br />
phylum Apicomplexa, supporting the hypothesis of a<br />
common molecular mechanism for host recognition<br />
attachment and invasion. They contain a combination<br />
of adhesive motifs including thrombospondin, integrin,<br />
apple and EGF-like domains. Invasion is successfully<br />
achieved when the newly formed parasitophorous<br />
vacuole is sealed and at this point, the<br />
tight interaction between host cell receptors and parasite<br />
ligands must be disrupted. The proteolytic processing<br />
of micronemal proteins that occurs on the<br />
parasite surface could fulfil such a function.<br />
TgMIC4 carries six apple domains and binds<br />
to host cells<br />
S. Brecht, U. Jäkle<br />
TgMIC4 carries six apple domains, a signal peptide<br />
at the amino terminus and no apparent membranespanning<br />
domain. Apple domains are found in plasma<br />
coagulation factors, kallikrein and factor XI and are<br />
involved in highly specific protein-protein interactions.<br />
MIC4 is expressed in all invasive stages of the<br />
parasites, secreted at the time of invasion and proteolytically<br />
processed twice onto the parasite surface,<br />
after secretion by the micronemes. MIC4 binds efficiently<br />
to host cells and the adhesive properties map<br />
within the carboxy-terminal apple domain of the protein.<br />
Preliminary experiments showed that galactose<br />
inhibits specifically binding to host cells and we are<br />
currently characterizing further the lectin properties<br />
of MIC4 and attempting to identify the host cell<br />
receptor(s) by expressing heterologously the apple<br />
domain in P. pastoris.<br />
133
A novel family of secreted protein carrying<br />
EGF-like domains.<br />
M. Meißner and M. Reiss<br />
We have characterized a new family of T. gondii<br />
micronemal proteins containing various numbers of<br />
EGF-like domains (TgMIC6, TgMIC7 TgMIC8 and<br />
TgMIC9) and their deduced amino acids sequences<br />
predict type I transmembrane proteins. The short<br />
cytoplasmic tails of these proteins are conserved<br />
across the Apicomplexa.<br />
According to the capping model for host cell invasion<br />
by Apicomplexa, transmembrane proteins are<br />
anticipated to establish tight and specific interactions<br />
with host cell receptors through their extracellular<br />
adhesive domains. At the same time, their carboxyterminal<br />
domain shall promote a direct or indirect<br />
connection with actomyosin system of the parasite<br />
to transfer the mechanical force across the plasma<br />
membrane. The cytoplasmic domains of these MICs<br />
failed to interact with TgMyoA tail in the yeast<br />
“two hybrid” assay. We are pursuing a “two hybrid”<br />
screening with a T. gondii cDNA fusion library and<br />
biochemical approaches to identify partners interacting<br />
with the MIC cytoplasm domains.<br />
TgMIC6 functions as cargo receptor with two<br />
soluble microneme proteins<br />
M. Reiss, N. Viebig, in collaboration with J.F.<br />
Dubremetz (Institut Pasteur, Lille)<br />
TgMIC6 carries three EGF-like domains, a transmembrane<br />
spanning region and a short conserved<br />
cytoplasmic tail. The disruption of MIC6 gene by<br />
homologous recombination revealed that this gene is<br />
not essential for the survival of tachyzoites in culture.<br />
Interestingly, the sorting of two soluble micronemal<br />
134<br />
proteins is compromised by the absence of MIC6.<br />
MIC1 and MIC4 are mistargeted into the default<br />
pathway, which in T. gondii transits through the<br />
dense granules and ends in the parasitophorous vacuole.<br />
The distribution of other micronemal proteins<br />
such as the soluble MIC3 and the transmembrane<br />
MIC2 are not affected in the mic6-mutant. We<br />
have demonstrated genetically by complementing<br />
the mic6-mutant with truncated or chimeras versions<br />
of MIC6 that this protein interacts with MIC1 and<br />
MIC4 and serve as specific receptor for sorting into<br />
the micronemes. Biochemical evidence also supports<br />
the genetic data and lead to the conclusion that MIC1,<br />
MIC4 and MIC6 are building a complex in the early<br />
compartment of the secretory pathway, which is prerequisite<br />
for the accurate sorting of the two soluble<br />
MICs. Analyses of deletion mutants of MIC6 indicate<br />
that the targeting signal to the micronemes is<br />
localized within the cytoplasmic tail domain of the<br />
protein. Confirming these results, the cytoplasmic<br />
tail of MIC2 contains identical sorting signals including<br />
a conserved tyrosine based motif, which likely<br />
involves interaction with a member of the adaptor<br />
complexes. MIC4 behaves as a neutral cargo protein<br />
whereas MIC1 appears to be involved in a quality<br />
control mechanism. In mutant parasites where MIC1<br />
gene has been disrupted by homologous recombination,<br />
both MIC4 and MIC6 are retained in the ER and<br />
Golgi. After discharge by the micronemes, MIC6 is<br />
distributed at the surface of the parasite and is rapidly<br />
released from the plasma membrane by proteolytic<br />
cleavage upstream of the transmembrane spanning<br />
domain. The multiple processing events occurring<br />
on the parasite surface might play a key role in<br />
disrupting the tight interaction between parasite and<br />
host cell membranes once invasion is achieved.<br />
III. A controlled gene expression system<br />
M. Meißner<br />
An inducible control of individual gene expression is<br />
prerequisite to the study of essential processes such<br />
as host cell invasion by an obligate intracellular parasite.<br />
The straightforward strategy would be to import<br />
the tetracycline-repressor TetR or transactivator tTA.<br />
A modified synthetic gene coding for tTA and tetRep<br />
gene in which the codon usage is remodeled in favor<br />
of a more GC-rich content, was kindly provided by<br />
the group of Dr. Bujard. Both synthetic tetRep and<br />
tTA were faithfully expressed as stable transgenes<br />
in T. gondii tachyzoites. We confirmed by gel shift<br />
retardation assay that both proteins have the capacity<br />
to bind to tetO sequences and in a tetracycline<br />
dependent fashion. All reporter plasmids tested so far<br />
failed to measure transactivation by tTA. In contrast<br />
a 10-fold repression of reporter gene expression was<br />
observed in presence of tetRep using a tubulin promoter<br />
where two tetO sequences have been introduced<br />
at the site of transcriptional initiation. The<br />
present results suggest that the VP16 activating<br />
domain fails to interact with the parasite transcription<br />
machinery. In contrast, the repressor system might<br />
provide with a system to manipulate the level of<br />
gene expression in T. gondii in a controlled manner<br />
although optimization of the inducibility is still indispensable.<br />
External Funding<br />
During the period reported our research was supported<br />
by grants from the Deutsche Forschungsgemeinschaft<br />
DFG (SFB 544 “Kontrolle tropischer<br />
Infektionskrankheiten”, Schwerpunkt “<strong>Molekulare</strong><br />
Motoren”, Schwerpunkt “Voraussetzungen und<br />
molekulare Mechanismen <strong>der</strong> Persistenz von Para–<br />
siten im Wirt”, Graduiertenkolleg “Kontrolle <strong>der</strong> Genexpression<br />
in pathogenen Mikroorganismen”, Projekt-’Sachbeihilfen’)<br />
and from the Humboldt Foundation.<br />
PUBLICATIONS<br />
Soldati, D., Lassen, A., Dubremetz, J.F. and Boothroyd,<br />
J.C. (1998). Processing of Toxoplasma ROP1<br />
protein in nascent rhoptries. Mol. Biochem. Parasitol.<br />
96, 37-48.<br />
Striepen, B., He, C.Y., Matrajt, M., Soldati, D. and<br />
Roos, D.S. (1998). Expression, selection, and organellar<br />
targeting of the green fluorescent protein in<br />
Toxoplasma gondii. Mol. Biochem. Parasitol. 92,<br />
325-338.<br />
Soëte, M., Hettman, C. and Soldati, D. (1999). The<br />
importance of reverse genetics in determining gene<br />
function in apicomplexan parasites. Parasitol. 118,<br />
S53-61.<br />
Di Cristina, M., Ghouze, F., Kocken, C.H., Naitza,<br />
S., Cellini, P., Soldati, D., Thomas, A.W. and Crisanti,<br />
A. (1999). Transformed Toxoplasma gondii<br />
tachyzoites expressing the circumsporozoite protein<br />
of Plasmodium knowlesi elicit a specific immune<br />
response in rhesus monkeys. Infect. Immun. 67,<br />
1677-1682.<br />
Brecht, S., Erdhart, H., Soete, M. and Soldati, D.<br />
(1999). Genome engineering of Toxoplasma gondii<br />
using the site-specific recombinase Cre. Gene 234,<br />
239-247.<br />
Jomaa, H., Wiesner, J., San<strong>der</strong>brand, S., Altincicek,<br />
135
B., Weidemeyer, C., Hintz, M., Turbachova, I., Eberl,<br />
M., Zeidler, J., Lichtenthaler, H.K. Soldati, D. and<br />
Beck, E. (1999). Inhibitors of the nonmevalonate<br />
pathway of isoprenoid biosynthesis as antimalarial<br />
drugs. Science 285, 1573-1576.<br />
Mattsson, J.G. and Soldati, D. (1999). MPS1: a small,<br />
evolutionarily conserved zinc finger protein from<br />
the protozoan Toxoplasma gondii. FEMS Microbiol.<br />
Lett. 180, 235-239.<br />
Hettmann, C. and Soldati, D. (1999). Cloning and<br />
analysis of a Toxoplasma gondii histone acetyltransferase:<br />
a novel chromatin remodelling factor<br />
in Apicomplexan parasites. Nucleic Acids Res. 27,<br />
4344-4352.<br />
Hettmann, C. Herm, A., Frank, B, Schwarz, E. Geiter,<br />
A. Soldati, T. and Soldati, D. (<strong>2000</strong>). A dibasic motif<br />
in the tail of a class XIV apicomplexan myosin is an<br />
essential determinant of plasma membrane localisation.<br />
Mol. Biol. Cell, 11, 1385-1400.<br />
Ding, M., Clayton C., and Soldati D. (<strong>2000</strong>). Toxoplasma<br />
gondii catalase: are there peroxisomes in<br />
Apicomplexa? J. Cell Sci., 113, 2409-2419.<br />
Di Cristina, M., Spaccapelo, R., Soldati, D., Bistoni,<br />
F., and Crisanti, A. (<strong>2000</strong>). Two conserved amino acid<br />
motifs mediate protein targeting to the micronemes of<br />
the apicomplexan parasite Toxoplasma gondii. Mol.<br />
Cell. Biol. (in press).<br />
Tomley, F. and Soldati, D. (<strong>2000</strong>). Mix and Match<br />
Modules: Structure and Function of Microneme Proteins<br />
in Apicomplexan Parasites. Parasitology Today<br />
(in press).<br />
136<br />
Ferguson, D.J.P., Brecht, S. and Soldati, D. (<strong>2000</strong>).<br />
The microneme protein MIC4, or an MIC4-like protein,<br />
is expressed within the macrogamete and associated<br />
with oocyst wall formation in Toxoplasma<br />
gondii. Int. J. Parasitol (in press).<br />
THESES<br />
Diploma<br />
Reiß, Matthias (1998). Identification and characterization<br />
of an invasion factor in Toxoplasma gondii.<br />
Geiter, Ariane (1998). Subcellular distribution of<br />
Toxoplasma gondii myosins.<br />
Ding, Martina (1999). Characterization of organelles<br />
in Toxoplasma gondii.<br />
Herm, Angelika (1999). Purification and characterization<br />
of recombinant myosins from Toxoplasma<br />
gondii.<br />
Viebig, Nicola (1999). Characterization of a sorting<br />
receptor for regulated soluble secretory proteins of<br />
Toxoplasma gondii.<br />
Sänger, Astrid (<strong>2000</strong>). Analysis and characterization<br />
of Toxoplasma gondii myosins B and C.<br />
Dissertations<br />
Brecht, Susan (<strong>2000</strong>). Characterization of a Toxoplasma<br />
gondii adhesin and establishment of Cre-lox<br />
recombinase system for T. gondii genome engineering.<br />
Hettmann, Christine (<strong>2000</strong>). Identification of a chromatin<br />
remodelling factor in Toxoplasma gondii and<br />
myosins as molecular motors for the invasion of host<br />
cells by the Apicomplexa.<br />
STRUCTURE OF THE GROUP<br />
E-mail: soldati@sun0.urz.uni-heidelberg.de<br />
Group lea<strong>der</strong> Soldati, Dominique, Dr.<br />
Postdoctoral<br />
fellows Soëte, Martine*<br />
Hernandez, Carmen *<br />
Delbac, Fre<strong>der</strong>ic*<br />
Ph.D. students Brecht, Susan, Dipl.-Biol.<br />
Hettmann, Christine, Dipl.-Biol.<br />
Reiss, Matthias, Dipl.-Biol *<br />
Meißner, Markus, Dipl.-Biol *<br />
Diploma students Geiter, Ariane*<br />
Herm, Angelika*<br />
Viebig, Nicola*<br />
Sänger, Astrid*<br />
Opitz, Corinna*<br />
Techn. assistant Jäkle, Ursula*<br />
* part of the time reported<br />
137
138<br />
Central Facilities<br />
Biomolecular Chemistry<br />
The chemical synthesis, structural analysis and the<br />
characterization of structure-activity relationships of<br />
biopolymers such as proteins and nucleic acids represent<br />
an important link between organic chemistry,<br />
physical chemistry and molecular biology. The chemical<br />
core facility operates in this interface region<br />
making biomolecular chemistry available to all scientists<br />
at the <strong>ZMBH</strong> and to numerous associated groups<br />
in the surrounding institutes. In particular for the following<br />
techniques instrumentation and practical support<br />
are available:<br />
- micropreparative isolation of proteins and peptides<br />
- synthesis and purification of peptides<br />
- DNA sequence analysis<br />
- mass spectrometrical analysis of biopolymers.<br />
In cooperation with biologists these techniques are<br />
continuously updated. That makes the newest methods<br />
available for the solution of biological problems.<br />
Similarly, the close proximity of members of the core<br />
facility and the biological research groups leads to frequent<br />
scientific exchange. Early interaction has proved<br />
to be very valuable.<br />
During the last years the market has changed consi<strong>der</strong>ably.<br />
Companies are now offering services for the synthesis<br />
of oligonucleotides at low prices. Therefore, we<br />
discontinued the synthesis of oligonucleotides since<br />
we could not compete with these specialized companies.<br />
We rather direct now our main focus on the analysis<br />
of proteins and the synthesis of peptides. The still<br />
growing number of complete sequences of procaryotic<br />
and eukaryotic genomes results in an increasing<br />
demand for protein technology. The catchword is<br />
proteome analysis. The facility is equipped with a<br />
MALDI instrument (matrix assisted laser desorption<br />
ionisation) and an electrospray ionisation (ESI) ion<br />
trap mass spectrometer. This enables us to characterize<br />
proteins, which have been separated by one- or<br />
two-dimensional gel electrophoresis and assign them<br />
to the corresponding genes.<br />
Although the majority of or<strong>der</strong>ed peptides is in the<br />
range of 10-20 amino acids, we are able to synthesize<br />
peptides longer than 100 amino acids (longest peptide<br />
115 amino acids) including certain additional chemical<br />
modifications.<br />
In parallel to the service function of the group several<br />
collaborative projects are pursued concerning the analysis<br />
of protein primary structure and protein function.<br />
These projects are supplemented by our studies on the<br />
chemistry and instrumentation for synthesis and analysis<br />
of biopolymers. Our goal is to provide the members<br />
of the institute with state of the art technology.<br />
Richard Herrmann<br />
139
Biocomputing<br />
Computing and communication via the internet are<br />
absolutely essential for the mo<strong>der</strong>n biological Sciences.<br />
Accordingly, the <strong>ZMBH</strong> is equipped with a<br />
powerful computer network that comprises by now<br />
three central Macintosh Servers, ≈180 Macintoshs for<br />
endusers, two Sun workstations, 20 Windows com–<br />
puters and 20 printers that are connected with each<br />
other through an Ethernet and a Local talk system.<br />
A consi<strong>der</strong>able amount of computer-controlled equipment,<br />
such as microscopes, fluor- and phosphoimagers,<br />
oligonucleotide- and polypeptide synthesisers, as<br />
well as DNA-sequencing machines, are also integrated<br />
into the network.<br />
The <strong>ZMBH</strong> employs a team of three computer specialists,<br />
complemented by one or more students,<br />
who maintain and up-date the hardware and software<br />
and give a comprehensive support to all users. This<br />
includes, for example, administration of the network<br />
and servers, set up of computers according to the<br />
individual user‘s requirements and the creation of<br />
customised databases for sophisticated, user-specific<br />
sequence analysis with UNIX-based software. In addition,<br />
the computer group provides software training<br />
in small groups or on an individual basis for a broad<br />
range of applications.<br />
140<br />
Raphael Mosbach<br />
Animal House and Transgene Lab 1<br />
The <strong>ZMBH</strong> animal house is a central division of the<br />
institute that provides research groups with the opportunity<br />
to breed experimental animals and perform<br />
animal experiments. For this purpose, the animal house<br />
maintains quantities of mice, rats, rabbits and African<br />
clawed toads (Xenopus laevis).<br />
Covering a total area of 560 m 2 there are rooms for<br />
housing the animals, preparation rooms, storage areas<br />
and a large enclosure with cage-washing facilities and<br />
autoclave. The animal house is run as a SPF-unit. That<br />
is to say, the animals are kept un<strong>der</strong> conditions that are<br />
strictly isolated from the outside environment. Careful<br />
controls ensure that animals introduced into the facility<br />
are free of any of the pathogens typically found in<br />
the species.<br />
Experimental protocols indicating immunizations,<br />
blood samplings etc. are performed by the animal<br />
house staff as a service to the research groups. Methods<br />
employed, for example, for treatment applications,<br />
body fluid samplings and surgery, adhere to contemporary<br />
standards in experimental animal science. Moreover<br />
the methods used are un<strong>der</strong> continuous assessment<br />
and improved techniques are introduced whenever<br />
possible.<br />
The Transgene-Laboratory offers the generation of<br />
transgenic mice and rats and mouse-chimeras, as well<br />
as offering in-vitro-fertilization, ovary-transfer and<br />
cryopreservation of embryos of both species. During<br />
1998/99 224 pronucleus- and ES-cell-injections were<br />
performed with mice, which delivered a total of over<br />
1.300 foun<strong>der</strong>s and chimeras respectively - an average<br />
of 6 positive animals per experiment. Also, 49<br />
transgenic mouse lines were cryopreserved, and 2 lines<br />
were revitalized. Finally, 2 invitro-fertilizations were<br />
performed and 11 microbilogically contaminated lines<br />
from other facilities were re<strong>der</strong>ived by embryotransfer.<br />
Moreover, courses on transgenic techniques and reproductive<br />
biology were held for graduate students, and<br />
special training programs were offered for transgene<br />
technicians.<br />
1 For more information use the link „Biogroup“ of the <strong>ZMBH</strong>-homepage<br />
Jürgen Weiß<br />
Teaching Facilities<br />
The <strong>ZMBH</strong> has invested consi<strong>der</strong>able funds and effort<br />
in building and maintaining adequate facilities for<br />
student laboratory courses in molecular biology, cell<br />
biology, biochemistry and microbiology. This teaching<br />
facility is organized as an independent unit consisting<br />
of a laboratory providing space for 14 students,<br />
with labs for special equipment, a darkroom and a<br />
kitchen for cleaning glassware and for media preparation.<br />
Major equipment includes: scintillation counters,<br />
spectrophotometers, ultracentrifuges and computers.<br />
In addition, there is all the equipment required<br />
for work with radioactive compounds and biological<br />
materials at the S1 safety level. The facilities are<br />
scheduled each year for the 10 months of the regular<br />
curriculum of the Faculty of Biology. For the remaining<br />
time, the laboratory is used for postgraduate training.<br />
Our teaching secretariat provides administrative support<br />
to all the teaching activities and to the development<br />
of curricula. It also keeps records of all <strong>ZMBH</strong><br />
students.<br />
In the academic year 1998/99 we had some 80 graduates<br />
and around 300 un<strong>der</strong>graduates participating in<br />
the institute‘s programs in molecular biology and cell<br />
biology. These two programs have been chosen by<br />
70% of the biology students after their „Vordiplom“ in<br />
1998/1999.<br />
Hans Peter Blaschkowski<br />
Richard Herrmann<br />
141
Library<br />
The library of the <strong>ZMBH</strong> offers a number of services<br />
to support the research groups of the <strong>ZMBH</strong>. Subscriptions<br />
to 43 scientific journals and 23 monographic<br />
series are being entertained and the access to<br />
online journals has been increased to 193 journals via<br />
the central library of the university. While the online<br />
journals can be accessed from every lab in the institute,<br />
two computers within the library allow in addition<br />
to use our collection of CD-ROMs which include<br />
e. g. „Current Protocols in Molecular Biology“ and<br />
„Current Protocols in Protein Science“. Finally, access<br />
to articles from way out journals or backissues is provided<br />
via the University‘s copy service.<br />
Besides all the hard- and software support, the library<br />
is a place for quiet and undisturbed study away from<br />
the sometimes hectic activity at the bench.<br />
142<br />
Hermann Bujard<br />
Documentation Service<br />
The <strong>ZMBH</strong> documentation service supply all type of<br />
visual communication supports. This service includes<br />
computer graphics, photography and graphic arts activities.<br />
The service provides drawings and figures destined<br />
for publications, poster sessions or conferences. The<br />
visual supports are produced as black and white or<br />
colour prints, as overhead transparencies or as slides.<br />
The computerized equipment of the service is based<br />
on two Apple Computers connected to a desktop colour<br />
scanner, a slide scanner, a 35 mm film exposure<br />
unit and InkJet photo quality colour printers A4 to<br />
A2 size. Software used are Canvas and Illustrator<br />
for drawing, Photoshop for photographic touchingup,<br />
QuarkXPress for DTP work and PowerPoint for<br />
slide production.<br />
Scientific photography is increasingly handled by<br />
computerized processes-which give high quality results<br />
in shorter times. For those who want to learn the<br />
use of these new imaging methods, the documentation<br />
service provides advice and courses.<br />
Finally other products of this service lab include any<br />
kind of photographic shots during <strong>ZMBH</strong> symposia<br />
and the desktop publishing and layout of the annual<br />
report as well as other documents that the <strong>ZMBH</strong><br />
edits.<br />
Yves Cully<br />
Central Administration<br />
The management of all personnel, of finances and of<br />
various administrative and technical facilities of the<br />
<strong>ZMBH</strong> lies within the responsibilities of our institute‘s<br />
central administration.<br />
During 1998/99, about 250 individuals have been<br />
working at the <strong>ZMBH</strong>. They include approx. 70 Ph.D.<br />
students, approx. 40 postdoctoral scientists and scholarship<br />
hol<strong>der</strong>s from 15 nations. In addition we have 3<br />
foreign group-lea<strong>der</strong>s and many guest scientists from<br />
foreign countries. As usual, we had a high turnover<br />
rate of approx. 100 per year.<br />
The <strong>ZMBH</strong>‘s financial structure is quite complex. The<br />
state of Baden-Württemberg has, for the past years,<br />
been the only basic financial supporter of the institute,<br />
which has allowed an effective and efficient management<br />
of the <strong>ZMBH</strong>‘s financial resources. However,<br />
the institute has also been affected by the drastic<br />
financial cuts in the public sector. In real terms, this<br />
means the loss of 5 staff positions until 2002 in the<br />
central infrastructure. Such personnel reductions can<br />
only be made by restructuring the work profile in the<br />
central services for maximum efficiency. To compensate<br />
for the job cuts, the <strong>ZMBH</strong> has been given the<br />
assurance of basic financial support for the coming<br />
years.<br />
From 1998-1999, the development of financial support<br />
outside the public sector corresponded to the number<br />
of occupied research group lea<strong>der</strong> positions.Due to the<br />
high staff fluctuation on this level as well as the complicated<br />
proceedings in the appointment of a professorship,<br />
there has been a noticeable drop in external<br />
funding for this period as compared with the previous<br />
period from 1996-1997. This trend is expected to even<br />
out when all the vacant research group lea<strong>der</strong> positions<br />
are filled.<br />
In 1998, the central administration introduced a data<br />
base system in the areas of finance and personnel.<br />
With this system, all research group lea<strong>der</strong>s, secretaries<br />
as well as staff members holding key functions<br />
are able to obtain in varying detail important financial<br />
and personnel information online. The next step is the<br />
coordination of this system with the SAP-data base<br />
system used by the main administration of the University<br />
of Heidelberg. The setting up and maintenance of<br />
the data base at the <strong>ZMBH</strong> plays a central role in<br />
the <strong>ZMBH</strong> administration by optimising and linking<br />
the administrative tasks as well as the wide range of<br />
support services available to the staff members for<br />
research and teaching.<br />
Jürgen Auer<br />
143
Electrical and Mechanical Workshop<br />
Experimental research requires functional equipment<br />
and technical facilities around the clock. The main<br />
task of the workshop is, therefore, the maintenance<br />
and repair of instruments and technical help in emergencies,<br />
whether it be the breakdown of an autoclave,<br />
a cold room, a freezer or a laminar flow hood. Custom-made<br />
equipment is another focus of the workshop‘s<br />
services, for example, electrophoretic equipment<br />
in the „<strong>ZMBH</strong>-design“, produced in a limited<br />
series, relieves some strain on the budgets of research<br />
groups. In the past two years, the workshop has fulfilled<br />
requests for about 1500 repair jobs, 700 emergency<br />
calls and has produced around 2500 pieces of<br />
equipment or parts thereof.<br />
144<br />
Matthias Pawlitschko<br />
Gert Stegmüller<br />
Head Technicians<br />
The <strong>ZMBH</strong> scientific staff includes two head technicians<br />
who support the research groups by un<strong>der</strong>taking<br />
organisational and technical tasks. Their responsibilities<br />
are partly assigned at the level of individual storeys,<br />
and partly towards the entire building; they also<br />
act as safety officers for all areas including radioactive<br />
and biological safety. Each of these scientists is supported<br />
by a technician.<br />
Tasks at „storey“ level are:<br />
- Discussing problems with individual research<br />
groups<br />
- Organisation of, and taking minutes for, floor<br />
meetings; implementing decisions<br />
- Administrating the floor budget<br />
- Purchase and maintenance of major communal<br />
equipment<br />
- Organisation of the washing-up kitchens<br />
- Central purchasing of certain dangerous materials<br />
such as radioactivity<br />
- Assisting in moving operations when groups arrive<br />
or depart<br />
Tasks for the whole <strong>ZMBH</strong> are:<br />
* Radioactive safety<br />
- Organisation and monitoring of the various special<br />
radioactive labs<br />
- Central disposal of waste<br />
- Regular radioactive safety courses for <strong>ZMBH</strong> employees<br />
* Biological safety<br />
- Making sure that the work is conducted in accordance<br />
with the biological safety laws<br />
- Giving advice to people writing biological safety<br />
applications<br />
- Regular biological safety courses for <strong>ZMBH</strong> employees<br />
* Other safety precautions<br />
* Disposal of chemical waste<br />
* Taking care of the central stores of spare apparatus<br />
Head Technician: Baumm, Axel<br />
1. and 2. floor (radioactive safety,<br />
other safety precautions)<br />
Assistant: Merx, Alexan<strong>der</strong><br />
Head Technician: Sawruk, Erich, Dr.<br />
3. and 4. floor (biological safety,<br />
chemical waste, central stores of<br />
spare apparatus)<br />
Assistant: Fabian, Gerd<br />
Axel Baumm<br />
Erich Sawruk<br />
145
146<br />
The Graduate Programme<br />
The graduate programme involves all diploma and<br />
PhD students at the <strong>ZMBH</strong>. The programme aims to<br />
promote contact among the graduate students and also<br />
between the scientific groups. A major goal is to expose<br />
and train the students to perform the many additional<br />
tasks expected from a scientist, like writing<br />
grant applications, preparing oral presentations, etc. In<br />
addition, talks, discussions and special method courses<br />
are offered by specialists from the house e.g. microscopic<br />
techniques, creation of transgenic animals, the<br />
use of software for the analysis of data and their presentation.<br />
The programme is organized by an elected committee<br />
of three students and two liason professors.<br />
At the beginning of their curriculum the graduate students<br />
obtain help to orientate themselves at the <strong>ZMBH</strong><br />
and to complete the necessary administration procedures.<br />
Additionally, organised parties and sport events<br />
serve to promote a sense of community.<br />
Weekend seminar in Schmitten<br />
Twice a year a small symposium organized by the<br />
graduate students takes place at a seminar centre in<br />
the Taunus. The students can develop their presentation<br />
skills by giving a research seminar in an informal<br />
setting.<br />
Our main philosophy is that science should be exciting<br />
and fun.<br />
147
<strong>ZMBH</strong>-Lehrprogramm im Grund- und Hauptstudium<br />
Studienjahr 1998/99<br />
Molekularbiologie und Zellbiologie im Grundstudium<br />
Grundvorlesung<br />
<strong>Biologie</strong> III, Teil Zellbiologie WS B. Dobberstein, D. Görlich,W. Huttner<br />
(16.1. - 13.2.98)<br />
Grundpraktika<br />
Grundpraktikum C Teil 3 SS R. Herrmann und Forschungsgruppenleiter<br />
(14 gleiche Wochenblöcke) des <strong>ZMBH</strong><br />
Seminare<br />
Einführung in das Studium <strong>der</strong> <strong>Biologie</strong> WS H. Bujard<br />
Einführung in das Studium <strong>der</strong> <strong>Biologie</strong> WS B. Dobberstein, P. Mayinger, C. Hölscher<br />
Einführung in das Studium <strong>der</strong> <strong>Biologie</strong> WS G. Multhaup, P. Prior<br />
Einführung in das Studium <strong>der</strong> <strong>Biologie</strong> WS R. Paro, G. Cavalli<br />
Einführung in die Neurowissenschaften SS K.-A. Nave<br />
148<br />
Molekularbiologie und Zellbiologie im Hauptstudium<br />
Vorlesungen<br />
Molekularbiologie I WS K. Beyreuther, H. Bujard<br />
Molekularbiologie II: Genetik, Entwicklungs- SS S. Freundlieb, R. Jansen, K.-A. Nave R. Paro<br />
biologie und Differenzierung<br />
Seminar zur Vorlesung Genetik, Entwicklungs- SS H. Bujard, R. Jansen, R. Paro<br />
biologie und Differenzierung<br />
Molekularbiologie III (Allgemeine Mikrobiologie) WS H.U. Schairer, B. Hauer, W. Plaga<br />
Seminar zur Vorlesung Molekularbiologie III WS H.U. Schairer, B. Hauer, W. Plaga<br />
(Allgemeine Mikrobiologie)<br />
Molekularbiologie IV: Virologie SS H. Schaller, U. Protzer, S. Urban<br />
Seminar zur Vorlesung Molekularbiologie IV SS H. Schaller, U. Protzer, S. Urban<br />
<strong>Molekulare</strong> Zellbiologie I WS B. Dobberstein, H. Herrmann-Lerdon, W.W.<br />
Franke, E. Hurt, D. Görlich<br />
Seminar begleitend zur Vorlesung WS B. Dobberstein, G. Layh-Schmitt,<br />
<strong>Molekulare</strong> Zellbiologie I D. Görlich, C. Harter<br />
Seminar begleitend zur Vorlesung: WS K. Simons, E. Hurt, H. Herrmann-Lerdon,<br />
<strong>Molekulare</strong> Zellbiologie U. Kutay<br />
Zellbiologie IV: <strong>Molekulare</strong> und Zelluläre SS W.B. Huttner zus. mit K. Beyreuther, M.<br />
Neurobiologie Brand, R. Brandt, H.-H. Gerdes, D. Langosch,<br />
K.-A. Nave, B. Sakmann, P. Seeburg,<br />
J. Trotter, K. Unsicher<br />
Transposon und DNA-Rekombination: WS D.-H. Lankenau, R. Paro<br />
Schrittmacher in <strong>der</strong> Evolution und Genetik<br />
Transposonbiologie: Begleitseminar zur Vorlesung WS D.-H. Lankenau, R. Paro<br />
Grundzüge <strong>der</strong> Immunologie SS P. Knolle, H. Schaller<br />
von Infektionskrankheiten<br />
149
Seminare<br />
Pathogene Mikroorganismen WS H. Schaller, G. Darai, S. Urban, U. Protzer<br />
Pathogene Mikroorganismen Bakterien, WS C. Clayton, D. Soldati, NN<br />
Eukaryonten<br />
Kontrolle <strong>der</strong> Genaktivität auf posttrans- WS C. Clayton, R.-P. Jansenkriptioneller<br />
Ebene<br />
<strong>Molekulare</strong> Mechanismen zellulärer WS R.-P. Jansen, F. Sauer, K.-A. Nave<br />
Differenzierung<br />
Genetische Grundlagen neurologischer WS K.-A. Nave, M. Sereda<br />
Erkrankungen<br />
Proteingene und ihre Funktion WS G. Multhaup, N.N.<br />
Mikrobielle Entwicklungs- und Regulations- SS H.U. Schairer, W. Plagamechanismen<br />
Mechanismen bakterieller Virulenz SS H.U. Schairer<br />
Mechanismen <strong>der</strong> Genregulation in Eukaryonten SS G. Merdes, R. Paro<br />
<strong>Biologie</strong> und Pathologie von Malariaparasiten und SS H. Bujard, H.-M. Müller<br />
neue Strategien zur Bekämpfung von Malaria tropica<br />
Mo<strong>der</strong>ne Methoden in <strong>der</strong> Proteinchemie SS G. Multhaup, L. Hesse<br />
Organell-Biogenese SS C. Clayton, D. Görlich<br />
Protein/Protein-Erkennung SS D. Langosch, S. Dübel, G. Multhaup, R.<br />
Brandt<br />
Neurobiologie des Lernens SS K.-A. Nave, A. Bartholomä, M. Rossner<br />
Entwicklungsneurobiologie SS W.B. Huttner, zusammen mit M. Brand, R.<br />
Brandt, R. Klein, K.-A. Nave, J. Trotter, K.<br />
Unsicker<br />
Zell-Regulation und Signaltransduktion SS B. Dobberstein, I. Hoffmann<br />
Mo<strong>der</strong>ne Methoden in <strong>der</strong> Proteinchemie SS G. Multhaup, D. Beher<br />
150<br />
Hauptpraktika<br />
Methoden <strong>der</strong> Molekularbiologie, 3-wöchig, WS R. Herrmann, G. Gounari, U. Kutay<br />
ganztägig<br />
Molekularbiologie A1, 3-wöchig, ganztägig WS K.-A. Nave, M. Rossner, D. Soldati<br />
Zellbiologie A1 WS Die Dozenten <strong>der</strong> Zellbiologie<br />
Molekularbiologie A2, 3-wöchig, ganztägig WS T. Hartmann, S. Lichtenthaler, K. Paliga, P.<br />
Prior, A. Weidemann, K. Beyreuther<br />
Molekularbiologie A2, 3-wöchig, ganztägig WS H. U. Schairer, W. Plaga, P. Prior, K.<br />
Willwand, K. Beyreuther<br />
<strong>Molekulare</strong> Untersuchungen <strong>der</strong> Drosophila WS R. Paro<br />
Entwicklung, 3-wöchig, ganztägig<br />
Begleitvorlesung zum Hauptpraktikum: <strong>Molekulare</strong> WS R. Paro<br />
Untersuchungen <strong>der</strong> Drosophila- Entwicklung,<br />
3-wöchig, ganztägig<br />
Molekularbiologie: Mechanismen <strong>der</strong> Transkriptions- SS H. Bujard, S. Freundlieb, H.-M. Müller und<br />
kontrolle, 3 Wochen, ganztägig Mitarbeiter<br />
Methoden <strong>der</strong> Virusdiagnostik, SS H. Schaller, P. Knolle, C. Kuhn, S. Urban<br />
3 Wochen, ganztägig,<br />
Molekularbiologie: Mikrobiologie, SS R. Herrmann, H. Schaller, B. Hauer,<br />
6 Wochen, ganztägig, C.H. Schrö<strong>der</strong>, H. Zentgraf<br />
Mikrobiologie <strong>für</strong> Nebenfächler und Lehramt SS R. Herrmann, C.H. Schrö<strong>der</strong>,<br />
3 Wochen, ganztägig,<br />
Molekularbiologie: Laborpraktika , Die Forschungsgruppenleiter des <strong>ZMBH</strong><br />
3-6 wöchig, ganztägig,<br />
Hauptpraktikum Zellbiologie: Proteintransport in SS B. Dobberstein, P. Mayinger, M. Pool,<br />
vivo und in vitro, 3 Wochen ganztägig M. Seedorf<br />
<strong>Molekulare</strong> Parasitologie, 3-wöchig, ganztägig. SS C. Clayton, D. Soldati, G. Layh-Schmitt, M.<br />
Lanzer<br />
Zellbiologie: Laborpraktika, Die Forschungsgruppenleiter des <strong>ZMBH</strong><br />
3-6 wöchig, ganztägig<br />
151
Studienprogramm <strong>für</strong> Graduierte des <strong>ZMBH</strong><br />
Wintersemester 1998/99<br />
<strong>ZMBH</strong>-Kolloquium C. Clayton und die Forschungsgruppenleiter des <strong>ZMBH</strong><br />
Cell Biology Lectures B. Dobberstein, F. Wieland<br />
(Graduiertenkolleg 230 und SFB 352)<br />
Arbeitsberichte <strong>für</strong> Mitglie<strong>der</strong> des C. Clayton und die an<strong>der</strong>en Dozenten des Grad.Kollegs<br />
Graduiertenkollegs Kontrolle <strong>der</strong> Genexpression<br />
bei pathogenen Organismen<br />
Seminare<br />
Regulation <strong>der</strong> Genaktivität H. Bujard<br />
Spezielle Arbeiten <strong>der</strong> Tropenmedizin H. Bujard<br />
Aktuelle Veröffentlichungen in <strong>der</strong> Entwick- R. Paro, F. Sauer<br />
lungsgenetik<br />
Neue Veröffentlichungen in <strong>der</strong> Zellbiologie B. Dobberstein, P. Mayinger, D. Görlich<br />
Neuere Arbeiten aus <strong>der</strong> Molekularbiologie H. Schaller, P. Knolle<br />
Literaturseminar H.U. Schairer, R. Herrmann<br />
Literaturseminar: RNA C. Clayton, R.-P. Jansen<br />
Neue Ergebnisse <strong>der</strong> Molekularbiologie M. Eilers, K.-A. Nave, C. Clayton<br />
Aktuelle Probleme <strong>der</strong> Molekular- und K. Beyreuther, G. Multhaup, T. Hartmann, P. Jäkälä,<br />
Neurobiologie K. Paliga P. Prior, S. Lichtenthaler, A. Weidemann<br />
Neuere Literatur <strong>der</strong> <strong>Molekulare</strong>n Neurobiologie K.-A. Nave<br />
Neuere Arbeiten <strong>der</strong> <strong>Molekulare</strong>n Zellbiologie H. Bujard, S. Freundlieb, D. Soldati<br />
<strong>Molekulare</strong> Entwicklungsbiologie R. Paro, F. Sauer<br />
Proteintransport B. Dobberstein und Mitarbeiter<br />
Intrazellulärer Proteintransport D. Görlich<br />
Hepatitis B Viren und Retroviren H. Schaller<br />
152<br />
Molekularbiologie mikrobieller Regulation H.U. Schairer, W. Plaga<br />
und Entwicklung<br />
Mitarbeiterseminar R. Herrmann<br />
<strong>Molekulare</strong> <strong>Biologie</strong> und Zellbiologie von C. Clayton<br />
Trypanosomen<br />
<strong>Molekulare</strong> Parasitologie C. Clayton, D. Soldati<br />
<strong>Molekulare</strong> Motoren D. Soldati, R.P. Jansen, T. Soldati, M. Manstein<br />
Molekularbiologie und Genetik <strong>der</strong> K. Beyreuther, G. Multhaup, T. Hartmann, P. Jäkälä,<br />
Alzheimer Krankheit K. Paliga, P. Prior, S. Lichtenthaler, A. Weidemann<br />
Neurogenetik K.-A. Nave, A. Bartholomä, M. Rossner, E.-M. Krämer<br />
Mechanismen <strong>der</strong> Genexpression F. Sauer<br />
Messenger RNA-Lokalisierung R.-P. Jansen<br />
Methodenkurse<br />
Sequenzanalyse Kurs 1: LASERGENE B. Reiner<br />
Sequenzanalyse Kurs 2: Internet, World Wide Web B. Reiner<br />
Literatursuche und -verarbeitung B. Reiner<br />
Dokumentationstechniken Y. Cully<br />
Grundlagen <strong>der</strong> Mikroskopie A. Baumm<br />
Immunisierung von Versuchstieren J. Weiss, R. Frank<br />
Herstellung transgener Mäuse und damit J. Weiss, F. Zimmermann, F. Sprengel<br />
verbundener Techniken<br />
153
Sommersemester 1999<br />
<strong>ZMBH</strong>-Kolloquium C. Clayton, H. Schaller und die Forschungsgruppenleiter<br />
des <strong>ZMBH</strong><br />
Neurobiology Lectures mit Gastrednern W.B. Huttner, R. Brandt, D. Langosch, K.-A. Nave und die<br />
an<strong>der</strong>en Forschungsgruppenleiter <strong>der</strong> Neurobiologie<br />
Kolloquium des Graduiertenkollegs <strong>Molekulare</strong> W.B. Huttner, K.-A. Nave und die an<strong>der</strong>en Dozenten des<br />
und zelluläre Neurobiologie mit Gastrednern Graduiertenkollegs Neurobiologie<br />
Methodische Weiterbildung <strong>für</strong> Mitglie<strong>der</strong> des W.B. Huttner, K.-A. Nave und die an<strong>der</strong>en Dozenten des<br />
Graduiertenkollegs <strong>Molekulare</strong> und zelluläre Graduiertenkollegs Neurobiologie<br />
Neurobiologie<br />
Seminare<br />
Regulation <strong>der</strong> Genaktivität H. Bujard, S. Freundlieb<br />
Spezielle Aspekte <strong>der</strong> Tropenmedizin H. Bujard, D. Soldati und Mitarbeiter<br />
Aktuelle Veröffentlichungen in <strong>der</strong> R. Paro, F. Sauer<br />
Entwicklungsgenetik<br />
Literaturseminar: Molecular cell biology B. Dobberstein und Mitarbeiter, D. Görlich<br />
Neuere Arbeiten aus <strong>der</strong> Molekularbiologie H. Schaller, P. Knolle<br />
Literaturseminar R. Herrmann, H.U. Schairer<br />
Aktuelle Probleme <strong>der</strong> Genetik, Molekular-, K. Beyreuther, G. Multhaup, T. Hartmann, C. Bergsdorf,<br />
Neuro- und Zellbiologie C. Bergmann, P. Jäkälä, K. Ishii, K. Paliga, P. Prior,<br />
A. Weidemann<br />
Neuere Arbeiten aus dem Gebiet <strong>der</strong> R. Herrmann, H.U. Schairer und Mitarbeiter<br />
biologischen Regulation<br />
Neuere Literatur <strong>der</strong> <strong>Molekulare</strong>n Neurobiologie K.-A. Nave<br />
Neue Ergebnisse <strong>der</strong> Forschungsarbeiten H. Bujard, S. Freundlieb und Mitarbeiter<br />
<strong>Molekulare</strong> Entwicklungsbiologie R. Paro, F. Sauer<br />
Hepatitis B Viren und Retroviren H. Schaller und Mitarbeiter<br />
Forschungsseminar: Protein targeting and sorting B. Dobberstein und Mitarbeiter<br />
Molekularbiologie von Myxobakterien H.U. Schairer und Mitarbeiter<br />
Mitarbeiterseminar R. Herrmann<br />
154<br />
Molekularbiologie und Zellbiologie bei C. Clayton<br />
Trypanosomen<br />
<strong>Molekulare</strong> Parasitologie C. Clayton, D. Soldati<br />
Neurogenetik K.-A. Nave<br />
Molekularbiologie und Genetik <strong>der</strong> K. Beyreuther, G. Multhaup, T. Hartmann, C. Bergsdorf,<br />
Alzheimer Krankheit SC. Bergmann, P. Jäkälä, K. Ishii, K. Paliga, P. Prior,<br />
A. Weidemann<br />
Methoden <strong>der</strong> Molekularbiologie und Genetik K. Beyreuther, G. Multhaup, T. Hartmann, C. Bergsdorf,<br />
<strong>der</strong> Alzheimer Krankheit C. Bergmann, P. Jäkälä, K. Ishii, K. Paliga, P. Prior,<br />
A. Weidemann<br />
Modelle neuronaler Entwicklung und Regeneration T. Hartmann, P. Jäkälä, C. Bergmann, K. Ishii, P. Prior,<br />
K. Beyreuther<br />
Proteingene bei <strong>der</strong> Alzheimer Krankheit G. Multhaup, L. Hesse, J. Kinter, C. Elle<br />
Methoden und Ergebnisse <strong>der</strong> molekularen D. Bohmann, M. Brand, M. Hassel, W. Huttner, T. Leitz,<br />
Entwicklungsbiologie B. Mechler, W. Müller, K.-A. Nave, C. Niehrs, R. Paro,<br />
E. Pollerberg, F. Sauer, G. Schütz<br />
Methodenkurse<br />
Sequenzanalyse Kurs 1: LASERGENE B. Reiner<br />
Sequenzanalyse Kurs 2: Internet, World Wide Web<br />
Literatursuche und -verarbeitung<br />
Dokumentation Y. Cully<br />
Grundlagen <strong>der</strong> Mikroskopie A. Baumm<br />
Herstellung transgener Tiere und damit J. Weiss, F. Zimmermann, S. Dlugosz<br />
zusammenhängende Techniken<br />
155
156<br />
Anhang / Appendix<br />
157
Z<strong>ZMBH</strong>-Colloquia, -Seminars and Cell<br />
Biology Lectures 1998/99 — Invited<br />
Speakers<br />
Acquati, F. (Milano, Italy)<br />
Gene hunting in the chromosome 21 Down Region at<br />
21q22.3<br />
Aktories, A. (Freiburg, Germany)<br />
Activation of Rho GTPases by bacterial protein<br />
toxins<br />
Allshire, R. (Edinburgh, UK)<br />
Utilising transcriptional silencing to dissect fission<br />
yeast centromere structure and function<br />
Asai, K. (Nara, Japan<br />
Disruption and transcription analysis of genes around<br />
oric in Bacillus subtilis<br />
Attardi, G. (Pasadena, USA)<br />
Genetic and functional thresholds in mitochondrial<br />
diseases: How much is enough?<br />
Bartenschlager, R. (Mainz, Germany)<br />
Molecular analysis of Hepatitis C virus replication<br />
Bauerle, R. (Basel, Schweiz)<br />
Strategies for regulation of metabolic pathways: the<br />
paradigm of differentially regulated isozymes in aromatic<br />
biosynthesis<br />
Berberich, C. (Medellin, Kolumbien)<br />
Strategies of control, diagnosis and vaccination against<br />
Leishmaniasis in an endemic region: The example<br />
Colombia, South America<br />
Bereswill, S. (Freiburg, Germany)<br />
Metal-dependent gene regulation in the stomach pathogen<br />
Helicobacter pylori<br />
158<br />
Bergeron, J.M. (Montreal, Canada)<br />
The role of calnexin in glycoprotein folding and quality<br />
control<br />
Berkhout, B. (Amsterdam, The Netherlands)<br />
Analysis of HIV-1 transcription with mutant/ revertant<br />
viruses<br />
Blumer, K. (München, Germany)<br />
Tierversuche - zum Wohle <strong>der</strong> Menschen? Ethische<br />
Aspekte zu einem kontroversen Thema<br />
Bonifacino, J. (Bethesda, USA)<br />
Signal-adaptor interactions in internalization and lysosomal<br />
targeting<br />
Brady, S.T. (Dallas, USA)<br />
Sculpting the functional architecture of the neuron:<br />
Myelinating glia and the axonal cytoskeleton<br />
Breakefield, X. (Charlestown, MA, USA)<br />
Early onset torsion dystonia: A nondegenerative neurologic<br />
disease caused by a dominant mutation in an<br />
HSP-like protein<br />
Brimacombe, R. (Berlin, Germany)<br />
Structure of ribosomal RNA at 13A resolution<br />
Bruss, V. (Göttingen, Germany)<br />
Generation and selection of Hepatitis B virus core<br />
gene mutations which block nucleocapsid envelopment<br />
Bukau, B. (Freiburg, Germany)<br />
The DnaK chaperone system<br />
Casaccia-Bonnefil, P. (New York, USA)<br />
The decision between survival and death in the oligodendroycte<br />
lineage<br />
Cattaneo, R. (Zürich, Switzerland)<br />
Towards therapeutic viruses based on a measles vaccine<br />
Chavrier, P. (Marseille, France)<br />
Function of Rho and ARF GTP-binding proteins in<br />
the regulation of actin cytoskeleton organization<br />
Conzelmann, K. (Tübingen, Germany)<br />
Rhabdoviruses: Targeted gene delivery and antivirals<br />
Cooper, J. A. (Brisbane, Australia)<br />
Mapping of conformational B cell epitopes within<br />
alpha-helical coiled coil proteins<br />
Delbac, F. (Aubiere, France)<br />
Immunological and molecular characterization of<br />
polar tube proteins in the microsporidia Encephalitozoon<br />
cuniculi<br />
Dikstein, R. (Jerusalem, Israel)<br />
TAF II 105 - a coactivator TFIID subunit involved in<br />
cytokine signaling and B cell specific transcription<br />
Dobbelstein, M. (Marburg, Germany)<br />
Adenoviral oncoproteins moonlighting as transporters<br />
Duboule, D. (Geneva, Switzerland)<br />
A genetic approach to vertebrate hox gene regulation<br />
Duesberg, P. (Mannheim, Germany)<br />
Is aneuploidy sufficient to cause cancer and genetic<br />
instability?<br />
Eaton, S. (Heidelberg, Germany)<br />
Planar polarization in Drosophila and vertebrate epithelia<br />
Endo, T. (Nagoya, Japan)<br />
Import and folding of mitochondrial proteins<br />
Erdmann, R. (Bochum, Germany)<br />
Components of the protein import machinery of peroxisomes<br />
Esser, A.F. (Kansas City, USA)<br />
The membrane attack complex of complement: or<br />
how to kill a bug without committing suicide<br />
Estevez, A.M. (Los Angeles, USA)<br />
Development of new tools for the study of RNA editing<br />
in Trypanosomes<br />
Gaul, U. (New York, USA)<br />
Establishment of neural connectivity in the developing<br />
visual system of Drosophila<br />
Gebhardt, R. (Leipzig, Germany)<br />
Positional expression of glutamine synthetase in the<br />
liver: features, regulation and implications<br />
Gorvel, J.-P. (Marseille, France)<br />
Membrane traffic of intracellular pathogens: The<br />
models of Brucella and Salmonella<br />
Grant, S. (Edinburgh, UK)<br />
The NMDA receptor - PSD95 multiprotein complex<br />
regulated by fyn: a central mechanism for synaptic<br />
plasticity and learning?<br />
Graumann, P. (Cambridge, USA)<br />
Arrangement and Active Segregation of Chromosomes<br />
in Bacteria<br />
Greber, U. (Zürich, Switzerland)<br />
Adenovirus entry: Transport of DNA Virus into the<br />
Cell and Viral DNA into the Nucleus<br />
Gripon, P. (Rennes, France)<br />
Effect of PreS1 deletions on L protein retention, translocation<br />
and viral assembly of HBV<br />
Gull, K. (Manchester, UK)<br />
Cell cycle, differentiation and cytoskeleton of African<br />
trypanosomes<br />
Hacker, J. (Würzburg, Germany)<br />
Pathogenicity islands and the evolution of infectious<br />
agents<br />
159
Hegemann, J. (Düsseldorf, Germany)<br />
The Saccharomyces cerevisiae genome - from structure<br />
to function<br />
Heise, T. (Hamburg, Germany)<br />
Control of Hepatitis B Virus RNA stability by host<br />
factors<br />
Hentze, M. (Heidelberg, Germany)<br />
Translational control by RNA binding proteins<br />
Herrlich, P. (Karlsruhe, Germany)<br />
Signalling goes RNA: Regulated alternative splicing<br />
of CD44<br />
Herz, J. (Dallas, USA)<br />
The LDL receptor gene family - from lipid metabolism<br />
to brain development<br />
Hoch, M. (Bonn, Germany)<br />
Control of organogenesis be Wingless and EGF receptor<br />
signaling at ecto<strong>der</strong>m/endo<strong>der</strong>m boundaries in the<br />
Drosophila gut<br />
Hoeijmakers, J. (Rotterdam, The Netherlands)<br />
DNA repair in mammals: from in vivo dynamics to<br />
human syndromes<br />
Hoey, T. (San Francisco, USA)<br />
Cytokine signalling during the immune response<br />
Hohn, B. (Basel, Switzerland)<br />
„Gene therapy“ in green eukaryotes<br />
Hopkins, C. (London, U.K.)<br />
Cytoplasmic coat mechanisms operating on the endocytotic<br />
pathways of fibroblasts and epithelial cells<br />
Horrocks, P. (Würzburg, Germany)<br />
Mutational analysis identifies a five base pair cis-acting<br />
sequence essential for GBP130 promoter activity<br />
in Plasmodium falciparum<br />
Hwang, J.-J. (Los Angeles, USA)<br />
Retroviral vectors for regulatable or tissue-specific<br />
160<br />
gene expression<br />
Jacobs, A. (Köln, Germany)<br />
Non-invasive imaging of gene expression by Positron-<br />
Emission-Tomography<br />
Jäckle, H. (Göttingen, Germany)<br />
Segmentation in Drosophila: From gradients to<br />
stripes<br />
Jentsch, S. (Martinsried, Germany)<br />
A novel ubiquitination factor, E4, is involved in multiubiquitin<br />
chain assembly<br />
Johnston, B. (Mountain View, CA, USA)<br />
RNA Padlocks: A new approach to gene inhibition<br />
Kafatos, F. (Heidelberg, Germany)<br />
Mosquito immunity and susceptibility of anopheles to<br />
parasites<br />
Kann, M. (Giessen, Germany)<br />
Nuclear Import of HBV Capsid and Genome<br />
Kennedy, M. (Pasadeny, USA)<br />
Signal transduction molecules at the postsynaptic density<br />
of CNS gutamatergic neurons<br />
Kioussis, D. (London, UK)<br />
Position effect variegation, chromatin structure and<br />
lineage commitment<br />
Kirchhoff, F. (Berlin, Germany)<br />
Transgenic mouse models to study glial physiology<br />
Klinkert, M. (Rom, Italy)<br />
New approaches to anti-schistosomal therapy<br />
Koszinowski, U. (München, Germany)<br />
Viral proteins responsible for immune evasion: The<br />
gene m153 of mouse cytomegalovirus<br />
Langowski, J. (Heidelberg, Germany)<br />
Large-scale structure of DNA and chromatin - modelling<br />
and experiments<br />
Laskey, R. (London, U.K.)<br />
Cell cycle control of DNA replication in normal and<br />
neoplastic cells<br />
Leclerus, D. (Paris, France)<br />
The insecticidal toxins and the virulence factors of<br />
Bacillus thuringiensis<br />
Lee, J. E. (Denver, USA)<br />
Neuro D: a differentiation factor for neuronal and pancreatic<br />
beta cells<br />
Lefranc, M.-P. (Montpellier, France)<br />
The immuno genetics database (IMGT): a necessity<br />
for analysis of the B and T cells repertoire<br />
Lengauer, C. (Baltimore, MD, USA)<br />
Genetic Instability - the Achilles‘ Heel of Human<br />
Cancer<br />
Lind, M. (Uppsala, Sweden)<br />
Cellular iron homeostasis<br />
Lingelbach, K. (Marburg, Germany)<br />
Secretion in Plasmodium falciparum<br />
Lord, M. (Coventry, U.K.)<br />
Toxin entry into mammalian cells: Retrograde transport<br />
and membrane translocation<br />
Luini, A. (Chieti, Italy)<br />
The lipid machinery of membrane fission: The role<br />
of CtBP/BARS and the acylation of lyso-phosphatidic<br />
acid<br />
Mandelkow, E. (Hamburg, Germany)<br />
Tau protein: Phosphorylation, role in intracellular traffic,<br />
and implications for Alzheimer‘s disease<br />
Manson, M.D. (College Station, USA)<br />
Mechanisms of transmembrane signaling and receptor<br />
crosstalk in bacterial chemotaxis<br />
Masters, C. (Melbourne, Australia)<br />
Aß is more important than plaques<br />
McBride, J.S. (Edinburgh, UK)<br />
Human antibody responses to P. falciparum MSP1<br />
Mettenleiter, T.C. (Riems, Germany)<br />
Role of Viral Glycoproteins in Entry and Exit of<br />
Herpes Viruses<br />
Michaeli, S. (Rehovot, Israel)<br />
Novel small RNAs in trypanosomes<br />
Nover, L. (Frankfurt/M., Germany)<br />
Heat stress response: A concert of transcription factors<br />
and chaperones<br />
Pahl, H. (Freiburg, Germany)<br />
Signal transduction and virulence regulation by two<br />
component systems<br />
Peles, E. (Rehovot, Israel)<br />
CASPR in neuron-glia interactions<br />
Pieters, J. (Basel, Switzerland)<br />
A phagosomal coat protein involved in intracellular<br />
survival of Mycobacteria<br />
Pines, J. (Cambridge, UK)<br />
4-dimensional control of mitosis<br />
Plückthun, A. (Zürich, Switzerland)<br />
From molecular interaction screening to directed<br />
molecular evolution<br />
Pohlmeyer, K. (Kiel, Germany)<br />
Solute Transport in Chloroplast<br />
Prunel, B. (Washington, USA)<br />
Science, more than a journal<br />
Quijada, L. (Madrid, Spain)<br />
Regulation of hsp70 gene expression in Leishmania<br />
infantum<br />
Reth, M. (Freiburg, Germany)<br />
Signal transduction from the B-cell antigen receptor<br />
161
Riekinen, P. Jr. (Kuopio, Finland)<br />
Effects of estrogen therapy on memory and Abeta<br />
levels in APP*Ps1 transgenic mice<br />
Robinson, C. (Coventry, U.K.)<br />
Novel pathways for the targeting of proteins in chloroplasts<br />
and bacteria<br />
Römisch, K. (London, U.K.)<br />
Export of protein from the ER lumen to the cytosol<br />
Rohlmann, A. (Göttingen, Germany)<br />
Tissue-specific k.o. of LRP (low-density lipoprotein<br />
receptor-related protein) in liver and brain of mice<br />
using the cre/loxP recombination system<br />
Roos, D.S. (Philadelphia, USA)<br />
Exploring eukaryotic cell biology via the molecular<br />
genetics of Toxoplasma gondii<br />
Rothblatt, J. (Columbia, USA)<br />
Extending the lifespan of C. elegans: What is needed<br />
to live longer?<br />
Sakar, M. (Manchester, U.K.)<br />
Analysis of the cell cycle and differentiation in African<br />
trypanosomes<br />
Saedler, H. (Köln, Germany)<br />
MADS box genes in flower development and evolution<br />
Schibler, U. (Geneva, Switzerland)<br />
Circadian timing in animals and cells<br />
Schliwa, M. (München, Germany)<br />
Organelle movements and motor proteins<br />
Schübeler, D. (Braunschweig, Germany)<br />
Site-specific recombination to express transgenes and<br />
to analyze Cis-acting sequences<br />
Schultz, U. (Freiburg, Germany)<br />
Interferons in HBV infection - studies in the duck<br />
model system<br />
162<br />
Shahi, S. (Dummersdorf, Germany)<br />
Detection of polymorphism in the gene of adipogenic<br />
transcription factor PPAR gamma: Peroxisome proliferator<br />
activated receptor gamma<br />
Silver, P.A. (Harvard, USA)<br />
Movement of macromolecules in and out of the<br />
nucleus<br />
Sinning, I. (Heidelberg, Germany)<br />
Role of SRP GTPases in protein transport<br />
Skoff, R. (Wane State Univ., USA)<br />
Central nervous system myelin protein genes are abundantly<br />
expressed in non-neural cell types including<br />
preimplantation embryos<br />
Smith, J.C. (Heidelberg, Germany)<br />
From sequence to function via structure: Molecular<br />
modelling of proteins, complexes and conformational<br />
change<br />
Spasic, M. (Belgrade, Yugoslavia)<br />
Maturation of penicillin G amidase of Providencia<br />
rettgeri<br />
Speranca, M.A. (Sao Paulo, Brazil)<br />
Construction and use of cdc2-chimeras to investigate<br />
cell cycle control in the human malaria parasite Plasmodium<br />
vivax<br />
Springer, S. (Berkeley, USA)<br />
Specific packaging of membrane proteins into COPII<br />
ER to Golgi transport vesicles<br />
StJohnston, D. (Cambridge, UK)<br />
mRNA localisation and axis formation in Drosophila<br />
Subramani, S. (San Diego, USA)<br />
Mechanisms of peroxisomal protein import and biogenesis<br />
Tafel, A. (Warschau, Poland)<br />
The domain structural of mitochondrial ribosomal<br />
protein NAM9 in yeast<br />
Tamkun, J. (Santa Cruz, USA)<br />
Chromatin remodeling factors and the control of cell<br />
fate in Drosophila<br />
Thoenen, H. (Martinsried, Germany)<br />
Modulatory role of neurotrophins in activity-mediated<br />
neuronal plasticity<br />
Van Meer, G. (Amsterdam, The Netherlands)<br />
Membrane lipid diversity in search of cellular functions<br />
Verma, I. (La Jolla, CA, USA)<br />
Gene Therapy: Problems and Prospects<br />
Vertel, B. (Chicago, USA)<br />
Biosynthesis of cartilage extracellular matrix molecules:<br />
Expected and unexpected pathways<br />
Voigt, H. (Lausanne, Switzerland)<br />
The role of myosin IB in Entamoeba histolytica<br />
Yoshida, K.-C. (Hiroshima, Japan)<br />
Reverse genetics after the whole genome sequencing:<br />
identification of a divergon involved in inositol catabolism<br />
in Bacillus subtilis and its regulation<br />
Watts, C. (Dundee, U.K.)<br />
Capture and processing of antigens for presentation<br />
on MHC molecules<br />
Way, M. (Heidelberg, Germany)<br />
Actin and its role in cell motility<br />
Wimmer, E. (New York, USA)<br />
Molecular Mechanisms of Poliovirus Pathogenesis:<br />
Can Poliovirus be Harnessed for Treatment of Brain<br />
Tumors?<br />
Wimmer, E.A. (Bayreuth, Germany)<br />
Bicoid-independent formation of thoracic segments<br />
in Drosophila and universal insect transgenesis<br />
Wintersberger, U. (Wien, Austria)<br />
Ribonucleases H of Saccharomyces cerevisiae and<br />
their evolutionary relatives from other organisms<br />
Zerial, M. (Heidelberg, Germany)<br />
Rab proteins in the biogenesis and function of endosomes<br />
163
Administrative and Technical Staff (31.12.1999)<br />
Labor <strong>für</strong> Biomole- N.N. Sekretariate/ Baro, Ina<br />
kulare Chemie Bosserhoff, Armin Secretary Coutinho, Karin<br />
Ellis, Margrit Demuth, Heidemarie<br />
Hudak, Manuela Güth-Köhler, Gerlind<br />
Rami, Jutta<br />
EDV/Biocomputing Mosbach, Raphael, Dipl.Phys. Reinig, Sibylle<br />
Reiner, Berta, Dipl.Math. Schmid, Ingrid<br />
Wingert, Winfried Schwarz, Annette<br />
Wee, Gaik-Pin<br />
Dokumentation Cully, Yves-Michel<br />
Spül-, Medien- Barcinski, Barbara<br />
Versuchstierhaltung/ Weiß, Jürgen, Dr. Küchen/Kitchens Dölling, Kerstin<br />
Transgenlabor Dlugosz, Sascha Dörich, Gerda<br />
Animal Facilities/ Fiala, Sandra Grimm, Martina<br />
Transgen Lab Gärtner, Ulrike Heil, Wilma<br />
Hartmann, Susanne Kienbacher, Ute<br />
Hühn, Martina Kobek, Martina<br />
Mees, Doris Meffert, Ingrid<br />
Michelberger, Yvonne Neuhäuser, Annerose<br />
Pfeffer, Margarita Veit, Gertrud<br />
Wayß, Sybille Weizenwieser, Christa<br />
Zimmermann, Frank Winkler, Helena<br />
Bibliothek/Library delValle, Christine Verwaltung/ Auer, Jürgen, Betriebs-<br />
Administration wirt (VWA)<br />
Lehreinheit/ Herrmann, Richard, Prof. Apfel, Roswitha<br />
Teaching unit Blaschkowski, H.P., Dr. Bier, Gabriela<br />
Kern, Andrea Kröner, Barbara<br />
Reiser, Walter, Dr. Wochlik, Claudia<br />
Zoffer, Roswitha<br />
Werkstätten/ Konrad, Michael<br />
Laborsicherheit, Baumm, Axel, Dipl. Biol. Hausmeister Mächtel, Christian<br />
Entsorgung Sawruk, Erich, Dr. Workshops/ Pawlitschko, Matthias<br />
Biol. Sicherheit/ Fabian, Gerd Caretaker Reis, Karl-Friedrich<br />
Lab safety Merx, Alexan<strong>der</strong> Stegmüller, Gerd<br />
164<br />
Budget 1999<br />
Haushalt des Landes / State Budget<br />
Personalmittel 8.994.850,00<br />
Sachmittel 2.017.700,00<br />
Investitionen 515.400,00<br />
Landeshaushalt gesamt/State Budget Total 11.527.950,00<br />
Drittmittel / Grant Monies<br />
Deutsche Forschungsgemeinschaft (DFG)<br />
SFB 352, SFB 317 und SFB 544 1.753.417,00<br />
Projektmittel aus Einzelför<strong>der</strong>ung 2.227.855,00<br />
Graduiertenkollegs 534.500,00<br />
Bundesministerium <strong>für</strong> Bildung, Wissenschaft,<br />
Forschung und Technologie (BMBF)<br />
Projektmittel aus Einzelför<strong>der</strong>ung einschl.<br />
Kooperationen im Rahmen <strong>der</strong> BioRegio 1.788.230,00<br />
Drittmittel überstaatlicher<br />
Organisationen (EU, HFSP u.a.) 291.023,00<br />
Projektmittel aus Einzelför<strong>der</strong>ung<br />
<strong>der</strong> Industrie 1.314.457,00<br />
Projektmittel aus Einzelför<strong>der</strong>ung<br />
sonstiger (VW-Stiftung etc.) 271.543,00<br />
Drittmittel gesamt/Grant Monies Total 8.181.025,00<br />
Gesamtbetrag des Budgets 1999 / Total 1999 19.708.975,00 DM<br />
Gesamtbetrag des Budgets 1999 / Total 1999 in EURO 10.077.038,90 EURO<br />
165