Nitrosopumilus maritimus genome reveals unique - de la Torre ...
Nitrosopumilus maritimus genome reveals unique - de la Torre ...
Nitrosopumilus maritimus genome reveals unique - de la Torre ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
0.5<br />
A.<br />
B.<br />
Thermop<strong>la</strong>sma volcanium GSS1 [rusticyanin; gi13542283]<br />
Acidithiobacillus ferrooxidans [rusticyanin; gi3282064]<br />
Thermobaculum terrenum ATCC BAA-798 [p<strong>la</strong>stocyanin; gi227373287]<br />
Sphaerobacter thermophilus DSM 20745 [p<strong>la</strong>stocyanin; gi229876615]<br />
Streptomyces sp. AA4 [hypothetical protein; gi256667890]<br />
Burkhol<strong>de</strong>ria cenocepacia PC184 [p<strong>la</strong>stocyanin; gi254250279]<br />
95<br />
Catenulispora acidiphi<strong>la</strong> DSM 44928 [blue (type 1) copper domain protein; gi256391720]<br />
Mycobacterium intracellu<strong>la</strong>re ATCC 13950 [hypothetical protein; gi254819204]<br />
83 Mycobacterium avium subsp. avium ATCC 25291 [copper-binding protein; gi254776845]<br />
99 Mycobacterium avium 104 [copper-binding protein; gi118462975]<br />
Nmar0747<br />
96 Nmar0361<br />
Candidatus Koribacter versatillis [p<strong>la</strong>stocyanin-like protein; gi94968815]<br />
Nmar0902<br />
Chromobacterium vio<strong>la</strong>ceum ATCC 12472 [hypothetical protein; gi34498952]<br />
Methylocel<strong>la</strong> silvestris BL2 [blue (type 1) copper domain protein; gi217976580]<br />
Methylobacterium nodu<strong>la</strong>ns ORS [blue (type 1) copper domain protein; gi220920817]<br />
Methylocel<strong>la</strong> silvestris BL2 [blue (type 1) copper domain protein; gi217976580]<br />
61 Nitrobacter hamburgensis X14 [blue (type 1) copper domain protein; gi92118183]<br />
Bradyrhizobium japonicum USDA 110 [putative Amicyanin precursor; gi27378126]<br />
Paracoccus <strong>de</strong>nitrificans PD1222 [amicyanin; gi119387457]<br />
Nmar0190<br />
Methanosarcina mazei Go1 [hypothetical protein; gi21226177]<br />
64 Methanosarcina acetivorans C2A [hypothetical protein; gi20090537]<br />
C. symbiosum A [copper-binding protein; gi118575216]<br />
99<br />
Nmar0167<br />
99 Nmar0621<br />
Methanosarcina mazei Go1 [copper-binding protein; gi21228445]<br />
Methanosarcina acetivorans C2A [p<strong>la</strong>stocyanin/azurin family copper-binding protein; gi20090218]<br />
62<br />
Methanosarcina mazei Go1 [copper-binding protein; gi21228446]<br />
100 Methanosarcina acetivorans C2A [p<strong>la</strong>stocyanin/azurin family copper-binding protein; gi20090219]<br />
Jannaschia sp. CCS1 [blue (type 1) copper domain protein; gi89056161]<br />
Nmar0718<br />
Thermobaculum terrenum ATCC BAA-798 [p<strong>la</strong>stocyanin; gi227373254]<br />
Deinococcus <strong>de</strong>serti VCD115 [copper-binding protein; gi226356528]<br />
Synechococcus sp. WH 8102 [p<strong>la</strong>stocyanin; gi33866031]<br />
91 Prochlorococcus marinus MIT 9312 [p<strong>la</strong>stocyanin; gi78778966]<br />
88<br />
Anabaena variabilisATCC 29413 [p<strong>la</strong>stocyanin; gi75908957]<br />
79<br />
Gloeobacter vio<strong>la</strong>ceus PCC 7421 [p<strong>la</strong>stocyanin; gi35212909]<br />
Methanococcus maripaludis C7 [copper-binding protein; gi150402166]<br />
82 Methanococcus maripaludis S2 [copper-binding protein; gi45358560]<br />
Nmar1913<br />
82<br />
Nmar1789<br />
Nmar0300<br />
Nmar0762<br />
C. symbiosum A [copper-binding protein; gi118576966]<br />
100 C. symbiosum A [copper-binding protein; gi118576965]<br />
Nmar0516<br />
Nmar1087<br />
C. symbiosum A [hypothetical protein; gi118575848]<br />
98 C. symbiosum A [hypothetical protein; gi118575831]<br />
C. symbiosum A [copper-binding protein; gi118575244]<br />
79<br />
Nmar0121<br />
uncultured marine Crenarchaeote [putative copper-binding protein; gi167045254]<br />
100 uncultured marine Crenarchaeote [putative copper-binding protein; gi167044521]<br />
100 uncultured marine Crenarchaeote [putative copper-binding protein; gi167042919]<br />
C. symbiosum A [copper-binding protein; gi118575724]<br />
100 Nmar0273<br />
Nmar0732<br />
Nmar0121<br />
Nmar0324<br />
79<br />
Nmar0167<br />
80<br />
C. symbiosum A [copper-binding protein; gi118576967]<br />
Nmar0190<br />
Nmar0273<br />
Nmar0300<br />
Nmar0324<br />
Nmar0361<br />
Nmar0516<br />
Nmar0621<br />
Nmar0718<br />
Nmar0732<br />
Nmar0747<br />
Nmar0762<br />
Nmar1087<br />
Nmar1789<br />
Nmar1913<br />
100 aa<br />
0.1<br />
Bacillus subtilis subsp. subtilis str. 168 [BdbD thiol-disulfi<strong>de</strong> oxidoreductase; gi 16080401]<br />
Bacillus amyloliquefaciens FZB42 [BdbD; gi 154687467]<br />
Paenibacillus <strong>la</strong>rvae subsp. <strong>la</strong>rvae BRL-230010 [disulfi<strong>de</strong> <strong>de</strong>hydrogenase D; gi 167461952]<br />
Deinococcus geothermalis DSM 11300 [DsbA oxidoreductase; gi 94984799]<br />
Thermobaculum terrenum ATCC BAA-798 [protein-disulfi<strong>de</strong> isomerase; gi 227374421]<br />
100<br />
C.<br />
100<br />
87<br />
82<br />
75<br />
Gemmatimonas aurantiaca T-27 [putative oxidoreductase; gi 226228008]<br />
Sphaerobacter thermophilus DSM 20745 [protein-disulfi<strong>de</strong> isomerase; gi 229876989]<br />
Thermomicrobium roseum DSM 5159 [DsbA oxidoreductase; gi 221635547]<br />
Thermus aquaticus Y51MC23 [DsbA oxidoreductase; gi 218295130]<br />
Thermobifida fusca YX [protein-disulfi<strong>de</strong> isomerase; gi 72161925]<br />
Mycobacterium kansasii ATCC 12478 [DsbA oxidoreductase; gi 240169146]<br />
Rubrobacter xy<strong>la</strong>nophilus DSM 9941 [DsbA oxidoreductase; gi 108804655]<br />
Streptomyces coelicolor A3(2) [hypothetical protein SCO5993; gi 21224330]<br />
100<br />
Streptomyces ghanaensis ATCC 14672 [hypothetical protein SghaA1_08748; gi 239928300]<br />
Symbiobacterium thermophilum IAM 14863 [hypothetical protein STH2058; gi 51893196]<br />
Roseiflexus castenholzii DSM 13941 [DsbA oxidoreductase; gi 156741642]<br />
100<br />
99<br />
100<br />
Chloroflexus aggregans DSM 9485 [DsbA oxidoreductase; gi 219849651]<br />
Chloroflexus aggregans DSM 9485 [DsbA oxidoreductase; gi 219847445]<br />
Marinobacter algico<strong>la</strong> DG893 [DsbA oxidoreductase; gi 149377658]<br />
Vibrio harveyi HY01 [DsbA oxidoreductase; gi 153832115]<br />
Vibrio parahaemolyticus RIMD 2210633 [hypothetical protein VPA0994; gi 28900849]<br />
Moritel<strong>la</strong> sp. PE36 [putative membrane protein; gi 149908466]<br />
99<br />
Shewanel<strong>la</strong> benthica KT99 [hypothetical protein KT99_10483; gi 163751466]<br />
Roseiflexus castenholzii DSM 13941 [protein-disulfi<strong>de</strong> isomerase-like protein; gi 156741356]<br />
75<br />
Roseiflexus castenholzii DSM 13941 [DsbA oxidoreductase; gi 156743646]<br />
Chloroflexus aurantiacus J-10-fl [DsbA oxidoreductase; gi 163848707]<br />
Herpetosiphon aurantiacus ATCC 23779 [DsbA oxidoreductase; gi 159896786]<br />
72<br />
100<br />
Chloroflexus aurantiacus J-10-fl [DsbA oxidoreductase; gi 163845898]<br />
Chloroflexus aggregans DSM 9485 [DsbA oxidoreductase; gi 219850456]<br />
Stigmatel<strong>la</strong> aurantiaca DW4/3-1 [conserved hypothetical protein; gi 115374845]<br />
Solibacter usitatus Ellin6076 [DsbA oxidoreductase; gi 116625220]<br />
Solibacter usitatus Ellin6076 [DsbA oxidoreductase; gi 116624599]<br />
82<br />
Syntrophobacter fumaroxidans MPOB [DsbA oxidoreductase; gi 116751066]<br />
Anaeromyxobacter <strong>de</strong>halogenans 2CP-1 [DsbA oxidoreductase; gi 220919173]<br />
100<br />
Myxococcus xanthus DK 1622 [putative lipoprotein; gi 108762810]<br />
Stigmatel<strong>la</strong> aurantiaca DW4/3-1 [disulfi<strong>de</strong> interchange protein; gi 115379912]<br />
Archaeoglobus fulgidus DSM 4304 [hypothetical protein AF1354; gi 11498950]<br />
Cenarchaeum symbiosum A [protein-disulfi<strong>de</strong> isomerase; gi 118575694]<br />
Nmar0752<br />
93<br />
91<br />
Nmar0179<br />
Nmar0218<br />
73<br />
79<br />
Nmar0740<br />
Nmar0175<br />
Natrialba magadii ATCC 43099 ]DsbA oxidoreductase; gi 224823141]<br />
Nmar1863<br />
90<br />
Cenarchaeum symbiosum A [protein-disulfi<strong>de</strong> isomerase; gi 118576454]<br />
Nmar1805<br />
67<br />
100<br />
Cenarchaeum symbiosum A [protein-disulfi<strong>de</strong> isomerase; gi 118576169]<br />
Nmar1606<br />
72<br />
63<br />
88<br />
Nmar0164<br />
Nmar0169<br />
Nmar1590<br />
Cenarchaeum symbiosum A [protein-disulfi<strong>de</strong> isomerase; gi 118575427]<br />
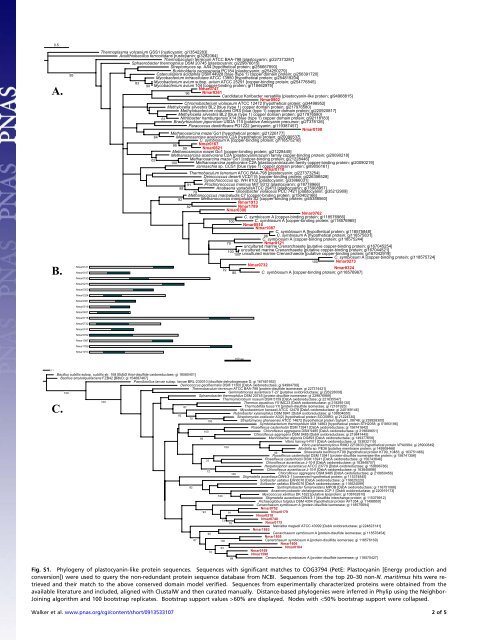
Fig. S1. Phylogeny of p<strong>la</strong>stocyanin-like protein sequences. Sequences with significant matches to COG3794 (PetE: P<strong>la</strong>stocyanin [Energy production and<br />
conversion]) were used to query the non-redundant protein sequence database from NCBI. Sequences from the top 20–30 non-N. <strong>maritimus</strong> hits were retrieved<br />
and their match to the above conserved domain mo<strong>de</strong>l verified. Sequences from experimentally characterized proteins were obtained from the<br />
avai<strong>la</strong>ble literature and inclu<strong>de</strong>d, aligned with ClustalW and then curated manually. Distance-based phylogenies were inferred in Phylip using the Neighbor-<br />
Joining algorithm and 100 bootstrap replicates. Bootstrap support values >60% are disp<strong>la</strong>yed. No<strong>de</strong>s with