Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
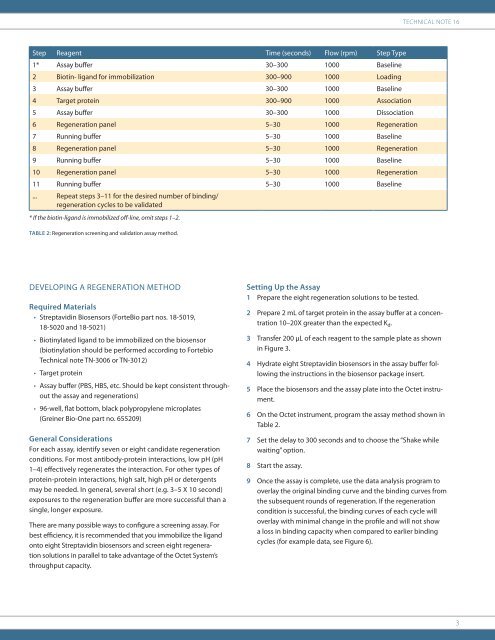
Technical Note 16Step Reagent Time (sec<strong>on</strong>ds) Flow (rpm) Step Type1* Assay buffer 30–300 1000 Baseline2 Biotin- ligand <str<strong>on</strong>g>for</str<strong>on</strong>g> immobilizati<strong>on</strong> 300–900 1000 Loading3 Assay buffer 30–300 1000 Baseline4 Target protein 300–900 1000 Associati<strong>on</strong>5 Assay buffer 30–300 1000 Dissociati<strong>on</strong>6 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>7 Running buffer 5–30 1000 Baseline8 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>9 Running buffer 5–30 1000 Baseline10 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>11 Running buffer 5–30 1000 Baseline... Repeat steps 3–11 <str<strong>on</strong>g>for</str<strong>on</strong>g> the desired number of binding/regenerati<strong>on</strong> cycles to be validated* If the biotin-ligand is immobilized off-line, omit steps 1–2.Table 2: <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> screening and validati<strong>on</strong> assay method.DEVELOPING A REGENERATION METHODRequired Materials• <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g> (<strong>ForteBio</strong> part nos. 18-5019,18-5020 and 18-5021)• Biotinylated ligand to be immobilized <strong>on</strong> the biosensor(biotinylati<strong>on</strong> should be per<str<strong>on</strong>g>for</str<strong>on</strong>g>med according to FortebioTechnical note TN-3006 or TN-3012)• Target protein• Assay buffer (PBS, HBS, etc. Should be kept c<strong>on</strong>sistent throughoutthe assay and regenerati<strong>on</strong>s)• 96-well, flat bottom, black polypropylene microplates(Greiner Bio-One part no. 655209)General C<strong>on</strong>siderati<strong>on</strong>sFor each assay, identify seven or eight candidate regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong>s. For most antibody-protein interacti<strong>on</strong>s, low pH (pH1–4) effectively regenerates the interacti<strong>on</strong>. For other types ofprotein-protein interacti<strong>on</strong>s, high salt, high pH or detergentsmay be needed. In general, several short (e.g. 3–5 X 10 sec<strong>on</strong>d)exposures to the regenerati<strong>on</strong> buffer are more successful than asingle, l<strong>on</strong>ger exposure.There are many possible ways to c<strong>on</strong>figure a screening assay. Forbest efficiency, it is recommended that you immobilize the ligand<strong>on</strong>to eight <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors and screen eight regenerati<strong>on</strong>soluti<strong>on</strong>s in parallel to take advantage of the Octet System’sthroughput capacity.Setting Up the Assay1 Prepare the eight regenerati<strong>on</strong> soluti<strong>on</strong>s to be tested.2 Prepare 2 mL of target protein in the assay buffer at a c<strong>on</strong>centrati<strong>on</strong>10–20X greater than the expected K d .3 Transfer 200 µL of each reagent to the sample plate as shownin Figure 3.4 Hydrate eight <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors in the assay buffer followingthe instructi<strong>on</strong>s in the biosensor package insert.5 Place the biosensors and the assay plate into the Octet instrument.6 On the Octet instrument, program the assay method shown inTable 2.7 Set the delay to 300 sec<strong>on</strong>ds and to choose the “Shake whilewaiting” opti<strong>on</strong>.8 Start the assay.9 Once the assay is complete, use the data analysis program tooverlay the original binding curve and the binding curves fromthe subsequent rounds of regenerati<strong>on</strong>. If the regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong> is successful, the binding curves of each cycle willoverlay with minimal change in the profile and will not showa loss in binding capacity when compared to earlier bindingcycles (<str<strong>on</strong>g>for</str<strong>on</strong>g> example data, see Figure 6).3