Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
Regeneration Strategies for Streptavidin Biosensors on ... - ForteBio
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
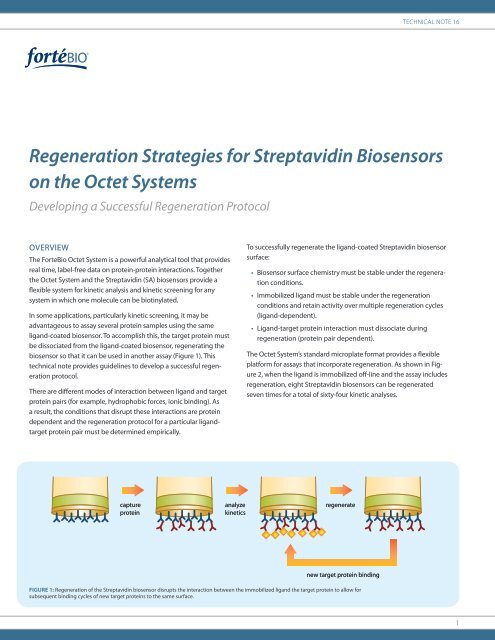
Technical Note 16<str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> <str<strong>on</strong>g>Strategies</str<strong>on</strong>g> <str<strong>on</strong>g>for</str<strong>on</strong>g> <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g><strong>on</strong> the Octet SystemsDeveloping a Successful <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> ProtocolOVERVIEWThe <strong>ForteBio</strong> Octet System is a powerful analytical tool that providesreal time, label-free data <strong>on</strong> protein-protein interacti<strong>on</strong>s. Togetherthe Octet System and the <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> (SA) biosensors provide aflexible system <str<strong>on</strong>g>for</str<strong>on</strong>g> kinetic analysis and kinetic screening <str<strong>on</strong>g>for</str<strong>on</strong>g> anysystem in which <strong>on</strong>e molecule can be biotinylated.In some applicati<strong>on</strong>s, particularly kinetic screening, it may beadvantageous to assay several protein samples using the sameligand-coated biosensor. To accomplish this, the target protein mustbe dissociated from the ligand-coated biosensor, regenerating thebiosensor so that it can be used in another assay (Figure 1). Thistechnical note provides guidelines to develop a successful regenerati<strong>on</strong>protocol.There are different modes of interacti<strong>on</strong> between ligand and targetprotein pairs (<str<strong>on</strong>g>for</str<strong>on</strong>g> example, hydrophobic <str<strong>on</strong>g>for</str<strong>on</strong>g>ces, i<strong>on</strong>ic binding). Asa result, the c<strong>on</strong>diti<strong>on</strong>s that disrupt these interacti<strong>on</strong>s are proteindependent and the regenerati<strong>on</strong> protocol <str<strong>on</strong>g>for</str<strong>on</strong>g> a particular ligandtargetprotein pair must be determined empirically.To successfully regenerate the ligand-coated <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensorsurface:• Biosensor surface chemistry must be stable under the regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong>s.• Immobilized ligand must be stable under the regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong>s and retain activity over multiple regenerati<strong>on</strong> cycles(ligand-dependent).• Ligand-target protein interacti<strong>on</strong> must dissociate duringregenerati<strong>on</strong> (protein pair dependent).The Octet System’s standard microplate <str<strong>on</strong>g>for</str<strong>on</strong>g>mat provides a flexibleplat<str<strong>on</strong>g>for</str<strong>on</strong>g>m <str<strong>on</strong>g>for</str<strong>on</strong>g> assays that incorporate regenerati<strong>on</strong>. As shown in Figure2, when the ligand is immobilized off-line and the assay includesregenerati<strong>on</strong>, eight <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors can be regeneratedseven times <str<strong>on</strong>g>for</str<strong>on</strong>g> a total of sixty-four kinetic analyses.captureproteinanalyzekineticsregeneratenew target protein bindingFigure 1: <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> of the <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensor disrupts the interacti<strong>on</strong> between the immobilized ligand the target protein to allow <str<strong>on</strong>g>for</str<strong>on</strong>g>subsequent binding cycles of new target proteins to the same surface.1
Technical Note 16Step Reagent Time (sec<strong>on</strong>ds) Flow (rpm) Step Type1* Assay buffer 30–300 1000 Baseline2 Biotin- ligand <str<strong>on</strong>g>for</str<strong>on</strong>g> immobilizati<strong>on</strong> 300–900 1000 Loading3 Assay buffer 30–300 1000 Baseline4 Target protein 300–900 1000 Associati<strong>on</strong>5 Assay buffer 30–300 1000 Dissociati<strong>on</strong>6 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>7 Running buffer 5–30 1000 Baseline8 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>9 Running buffer 5–30 1000 Baseline10 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 5–30 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>11 Running buffer 5–30 1000 Baseline... Repeat steps 3–11 <str<strong>on</strong>g>for</str<strong>on</strong>g> the desired number of binding/regenerati<strong>on</strong> cycles to be validated* If the biotin-ligand is immobilized off-line, omit steps 1–2.Table 2: <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> screening and validati<strong>on</strong> assay method.DEVELOPING A REGENERATION METHODRequired Materials• <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g> (<strong>ForteBio</strong> part nos. 18-5019,18-5020 and 18-5021)• Biotinylated ligand to be immobilized <strong>on</strong> the biosensor(biotinylati<strong>on</strong> should be per<str<strong>on</strong>g>for</str<strong>on</strong>g>med according to FortebioTechnical note TN-3006 or TN-3012)• Target protein• Assay buffer (PBS, HBS, etc. Should be kept c<strong>on</strong>sistent throughoutthe assay and regenerati<strong>on</strong>s)• 96-well, flat bottom, black polypropylene microplates(Greiner Bio-One part no. 655209)General C<strong>on</strong>siderati<strong>on</strong>sFor each assay, identify seven or eight candidate regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong>s. For most antibody-protein interacti<strong>on</strong>s, low pH (pH1–4) effectively regenerates the interacti<strong>on</strong>. For other types ofprotein-protein interacti<strong>on</strong>s, high salt, high pH or detergentsmay be needed. In general, several short (e.g. 3–5 X 10 sec<strong>on</strong>d)exposures to the regenerati<strong>on</strong> buffer are more successful than asingle, l<strong>on</strong>ger exposure.There are many possible ways to c<strong>on</strong>figure a screening assay. Forbest efficiency, it is recommended that you immobilize the ligand<strong>on</strong>to eight <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors and screen eight regenerati<strong>on</strong>soluti<strong>on</strong>s in parallel to take advantage of the Octet System’sthroughput capacity.Setting Up the Assay1 Prepare the eight regenerati<strong>on</strong> soluti<strong>on</strong>s to be tested.2 Prepare 2 mL of target protein in the assay buffer at a c<strong>on</strong>centrati<strong>on</strong>10–20X greater than the expected K d .3 Transfer 200 µL of each reagent to the sample plate as shownin Figure 3.4 Hydrate eight <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors in the assay buffer followingthe instructi<strong>on</strong>s in the biosensor package insert.5 Place the biosensors and the assay plate into the Octet instrument.6 On the Octet instrument, program the assay method shown inTable 2.7 Set the delay to 300 sec<strong>on</strong>ds and to choose the “Shake whilewaiting” opti<strong>on</strong>.8 Start the assay.9 Once the assay is complete, use the data analysis program tooverlay the original binding curve and the binding curves fromthe subsequent rounds of regenerati<strong>on</strong>. If the regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong> is successful, the binding curves of each cycle willoverlay with minimal change in the profile and will not showa loss in binding capacity when compared to earlier bindingcycles (<str<strong>on</strong>g>for</str<strong>on</strong>g> example data, see Figure 6).3
<str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> <str<strong>on</strong>g>Strategies</str<strong>on</strong>g> <str<strong>on</strong>g>for</str<strong>on</strong>g> <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g> <strong>on</strong> the Octet Systems12 3 4 5 6 7 8 9 10 11 12ABCDEFGHAssay BufferBiotin-proteinHuman IgG5 M NaCl0.01% SDSNaOH pH 10NaOH pH 11HCl, pH 0.510 mM Glycine, pH 110 mM Glycine, pH 2500 mM Phosphoric AcidFigure 4: Sample plate layout <str<strong>on</strong>g>for</str<strong>on</strong>g> the screening and validati<strong>on</strong> of up to 8different regenerati<strong>on</strong> reagents.EXAMPLE: DETERMINING AND VALIDATINGREGENERATION CONDITIONS FOR A RECEPTOR/PROTEIN INTERACTION ON STREPTAVIDIN BIOSENSORSIn this example, the optimal regenerati<strong>on</strong> c<strong>on</strong>diti<strong>on</strong>s <str<strong>on</strong>g>for</str<strong>on</strong>g> immobilizedbiotin-Protein A were determined. The objective was to identify thec<strong>on</strong>diti<strong>on</strong>s that enable the regenerati<strong>on</strong> of Protein A binding afterhIgG associati<strong>on</strong> <str<strong>on</strong>g>for</str<strong>on</strong>g> at least eleven binding/regenerati<strong>on</strong> cycles.Required Reagents• Assay Buffer (0.1 mg/mL BSA, 0.002% Tween-20, PBS)• 2 mL of 5 µg/mL stock of biotin-Protein A in Assay Buffer<str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> C<strong>on</strong>diti<strong>on</strong>s Tested• 5 M NaCl• 0.01% SDS• NaOH, pH 10• NaOH, pH 11• HCl, pH 0.5• 10 mM glycine, pH 1• 10 mM glycine, pH 2• 500 mM phosphoric acid• 2 mL of 66 nM soluti<strong>on</strong> of hIgG in Assay Buffer• 8 <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors hydrated in Assay BufferStep Reagent Time (sec) Flow (rpm) Step Type1 Assay Buffer 120 1000 Baseline2 5 µg/mL biotin Protein A 120 1000 Loading3 Assay Buffer 60 1000 Baseline4 hIgG, 66 nM 120 1000 Associati<strong>on</strong>5 Assay Buffer 120 1000 Dissociati<strong>on</strong>6 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 10 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>7 Assay Buffer 10 1000 Baseline8 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 10 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>9 Assay Buffer 10 1000 Baseline10 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> panel 10 1000 <str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g>11 Assay Buffer 60 1000 Baseline…Repeat steps 3-11 10 additi<strong>on</strong>al timesTable 3: Screening assay method <str<strong>on</strong>g>for</str<strong>on</strong>g> Protein A regenerati<strong>on</strong> c<strong>on</strong>diti<strong>on</strong>s.4
Technical Note 16nm5.004.504.003.503.002.502.001.501.000.500.000 200 400 600 800 1,000 1,200 1,400 1,600 1,800 2,000 2,200 2,400 2,600 2,800 3,000 3,200 3,400 3,600 3,800 4,000 4,200T ime (sec)A1 B1 C1 D1 E1 F1 G1 H1Figure 5: Real time results from regenerati<strong>on</strong> scouting and validati<strong>on</strong> experiment run <strong>on</strong> the Octet RED instrument. Data shown is from 8 <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors with biotinproteinA immobilized, binding human IgG and taken through regenerati<strong>on</strong> with 8 different reagents over 11 binding cycles..Assay SetupThe sample plate was set up as shown in Figure 4 and run <strong>on</strong> theOctet System using the assay method shown in Table 3.Assay ResultsIn Figure 5, the real time binding chart <str<strong>on</strong>g>for</str<strong>on</strong>g> the assay shows significantdifferences in the effectiveness of the eight regenerati<strong>on</strong>soluti<strong>on</strong>s across the eleven binding cycles tested. Only the acidicc<strong>on</strong>diti<strong>on</strong>s (sensors E1–H1) appear to show reas<strong>on</strong>able regenerati<strong>on</strong>of the binding capacity after the first binding cycle.Using the Data Analysis software, the successive binding cycles fromeach sensor can easily be overlaid (Figure 6A–H). The best regenerati<strong>on</strong>c<strong>on</strong>diti<strong>on</strong>s are quickly assessed by determining the level of IgGbinding <str<strong>on</strong>g>for</str<strong>on</strong>g> biosensors across all binding cycles. In this example, theNaCl, SDS and basic regenerati<strong>on</strong> soluti<strong>on</strong>s all show poor regenerati<strong>on</strong>as is evident by the clear loss of the amount of hIgG bindingABCDEnmnmFnmnmnmGnmnmHnmBindingCycle1234567891011Figure 6: Analysis of effectiveness of eight different reagents in regenerating Protein A immobilized to <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> biosensors. Reagents tested were: A) 5 M NaCl; B) 0.01%SDS; C) NaOH, pH 10; D) NaOH, pH 11; E) HCln pH 0.5; F) 10 mM Glycine pH 1; G) 10 mM glycine, pH 2; H) 500 mM phosphoric acid5
<str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> <str<strong>on</strong>g>Strategies</str<strong>on</strong>g> <str<strong>on</strong>g>for</str<strong>on</strong>g> <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g> <strong>on</strong> the Octet Systemsfrom binding cycle 1 to 11 (Figure 6A–D). C<strong>on</strong>diti<strong>on</strong>s HCl, glycinepH 1, glycine pH 2 and phosphoric acid all show good reproducibilityof the hIgG binding across all eleven cycles (Figure 6E–H). Inparticular, the 500 mM phosphoric acid (sensor H1) reagent showsvery good regenerati<strong>on</strong> of the protein A surface with a high level ofreproducibility of hIgG binding throughout the assay.From this single experiment a regenerati<strong>on</strong> c<strong>on</strong>diti<strong>on</strong> of 3X10sec<strong>on</strong>d cycles of 500 mM phosphoric acid has been identifiedas an excellent reagent to regenerate this receptor <strong>on</strong> <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g>biosensors.6
Technical Note 167
<str<strong>on</strong>g>Regenerati<strong>on</strong></str<strong>on</strong>g> <str<strong>on</strong>g>Strategies</str<strong>on</strong>g> <str<strong>on</strong>g>for</str<strong>on</strong>g> <str<strong>on</strong>g>Streptavidin</str<strong>on</strong>g> <str<strong>on</strong>g>Biosensors</str<strong>on</strong>g> <strong>on</strong> the Octet Systems8<strong>ForteBio</strong>, Inc.1360 Willow Road, Suite 201Menlo Park, CA 94025t: 888.OCTET-QK or 650.322.1360<strong>ForteBio</strong>, UK, Ltd.83 Victoria Street, Suite 407L<strong>on</strong>d<strong>on</strong>, SW1H 0HWUKt: +44-(0)20-31784425DistributorsVisit www.<str<strong>on</strong>g>for</str<strong>on</strong>g>tebio.com<str<strong>on</strong>g>for</str<strong>on</strong>g> a complete list of ourdistributors© 2009 <strong>ForteBio</strong>, Inc., <strong>ForteBio</strong>, and the<strong>ForteBio</strong> logo are trademarks and/orregistered trademarks of <strong>ForteBio</strong>, Inc.P/N TN-3016 Rev A