articlesTable 14 SSR content <strong>of</strong> <strong>the</strong> <strong>human</strong> <strong>genome</strong>Length <strong>of</strong> repeat unit Average bases per Mb Average number <strong>of</strong> SSRelements per Mb1 1,660 36.72 5,046 43.13 1,013 11.84 3,383 32.55 2,686 17.66 1,376 15.27 906 8.48 1,139 11.19 900 8.610 1,576 8.611 770 8.7.............................................................................................................................................................................SSRs were identi®ed by using <strong>the</strong> computer program T<strong>and</strong>em Repeat Finder with <strong>the</strong> followingparameters: match score 2, mismatch score 3, indel 5, minimum alignment 50, maximum repeatlength 500, minimum repeat length 1.tellites, whereas those with longer repeat units (n = 14±500 bases)are <strong>of</strong>ten termed minisatellites. With <strong>the</strong> exception <strong>of</strong> poly(A) tailsfrom reverse transcribed messages, SSRs are thought to arise byslippage during DNA replication 212,213 .We compiled a catalogue <strong>of</strong> all SSRs over a given length in <strong>the</strong><strong>human</strong> draft <strong>genome</strong> sequence, <strong>and</strong> studied <strong>the</strong>ir properties(Table 14). SSRs comprise about 3% <strong>of</strong> <strong>the</strong> <strong>human</strong> <strong>genome</strong>, with<strong>the</strong> greatest single contribution coming from dinucleotide repeats(0.5%). (The precise criteria for <strong>the</strong> number <strong>of</strong> repeat units <strong>and</strong> <strong>the</strong>extent <strong>of</strong> divergence allowed in an SSR affect <strong>the</strong> exact census, butnot <strong>the</strong> qualitative conclusions.)There is approximately one SSR per 2 kb (<strong>the</strong> number <strong>of</strong> nonoverlappingt<strong>and</strong>em repeats is 437 per Mb). The catalogue con®rmsvarious properties <strong>of</strong> SSRs that have been inferred from samplingapproaches (Table 15). The most frequent dinucleotide repeats areAC <strong>and</strong> AT (50 <strong>and</strong> 35% <strong>of</strong> dinucleotide repeats, respectively),whereas AG repeats (15%) are less frequent <strong>and</strong> GC repeats (0.1%)are greatly under-represented. The most frequent trinucleotides areAAT <strong>and</strong> AAC (33% <strong>and</strong> 21%, respectively), whereas ACC (4.0%),AGC (2.2%), ACT (1.4%) <strong>and</strong> ACG (0.1%) are relatively rare.Overall, trinucleotide SSRs are much less frequent than dinucleotideSSRs 214 .SSRs have been extremely important in <strong>human</strong> genetic studies,because <strong>the</strong>y show a high degree <strong>of</strong> length polymorphism in <strong>the</strong><strong>human</strong> population owing to frequent slippage by DNA polymeraseduring replication. Genetic markers based on SSRsÐparticularly(CA) n repeatsÐhave been <strong>the</strong> workhorse <strong>of</strong> most <strong>human</strong> diseasemappingstudies 101,102 . The availability <strong>of</strong> a comprehensive catalogue<strong>of</strong> SSRs is thus a boon for <strong>human</strong> genetic studies.The SSR catalogue also allowed us to resolve a mystery regardingmammalian genetic maps. Such genetic maps in rat, mouse <strong>and</strong><strong>human</strong> have a de®cit <strong>of</strong> polymorphic (CA) n repeats on chromosomeX 30,101 . There are two possible explanations for this de®cit. Theremay simply be fewer (CA) n repeats on chromosome X; or (CA) nrepeats may be as dense on chromosome X but less polymorphic in<strong>the</strong> population. In fact, <strong>analysis</strong> <strong>of</strong> <strong>the</strong> draft <strong>genome</strong> sequence showsthat chromosome X has <strong>the</strong> same density <strong>of</strong> (CA) n repeats per Mb as<strong>the</strong> autosomes (data not shown). Thus, <strong>the</strong> de®cit <strong>of</strong> polymorphicmarkers relative to autosomes results from population geneticforces. Possible explanations include that chromosome X has asmaller effective population size, experiences more frequent selectivesweeps reducing diversity (owing to its hemizygosity in males),or has a lower mutation rate (owing to its more frequent passagethrough <strong>the</strong> less mutagenic female germline). The availability <strong>of</strong> <strong>the</strong>draft <strong>genome</strong> sequence should provide ways to test <strong>the</strong>se alternativeexplanations.Segmental duplicationsA remarkable feature <strong>of</strong> <strong>the</strong> <strong>human</strong> <strong>genome</strong> is <strong>the</strong> segmentalduplication <strong>of</strong> portions <strong>of</strong> genomic sequence 215±217 . Such duplicationsinvolve <strong>the</strong> transfer <strong>of</strong> 1±200-kb blocks <strong>of</strong> genomic sequenceto one or more locations in <strong>the</strong> <strong>genome</strong>. The locations <strong>of</strong> bothdonor <strong>and</strong> recipient regions <strong>of</strong> <strong>the</strong> <strong>genome</strong> are <strong>of</strong>ten not t<strong>and</strong>emlyarranged, suggesting mechanisms o<strong>the</strong>r than unequal crossing-overfor <strong>the</strong>ir origin. They are relatively recent, inasmuch as strongsequence identity is seen in both exons <strong>and</strong> introns (in contrast toregions that are considered to show evidence <strong>of</strong> ancient duplications,characterized by similarities only in coding regions). Indeed,many such duplications appear to have arisen in very recentevolutionary time, as judged by high sequence identity <strong>and</strong> by<strong>the</strong>ir absence in closely related species.Segmental duplications can be divided into two categories. First,interchromosomal duplications are de®ned as segments that areduplicated among nonhomologous chromosomes. For example, a9.5-kb genomic segment <strong>of</strong> <strong>the</strong> adrenoleukodystrophy locus fromXq28 has been duplicated to regions near <strong>the</strong> centromeres <strong>of</strong>chromosomes 2, 10, 16 <strong>and</strong> 22 (refs 218, 219). Anecdotal observationssuggest that many interchromosomal duplications map near <strong>the</strong>centromeric <strong>and</strong> telomeric regions <strong>of</strong> <strong>human</strong> chromosomes 218±233 .The second category is intrachromosomal duplications, whichoccur within a particular chromosome or chromosomal arm. Thiscategory includes several duplicated segments, also known as lowcopy repeat sequences, that mediate recurrent chromosomal structuralrearrangements associated with genetic disease 215,217 . Exampleson chromosome 17 include three copies <strong>of</strong> a roughly 200-kb repeatseparated by around 5 Mb <strong>and</strong> two copies <strong>of</strong> a roughly 24-kb repeatseparated by 1.5 Mb. The copies are so similar (99% identity) thatparalogous recombination events can occur, giving rise to contiguousgene syndromes: Smith±Magenis syndrome <strong>and</strong> Charcot±Marie±Tooth syndrome 1A, respectively 34,234 . Several o<strong>the</strong>r examplesare known <strong>and</strong> are also suspected to be responsible for recurrentmicrodeletion syndromes (for example, Prader±Willi/Angelman,Table 15 SSRs by repeat unitRepeat unitNumber <strong>of</strong> SSRs per MbAC 27.7AT 19.4AG 8.2GC 0.1AAT 4.1AAC 2.6AGG 1.5AAG 1.4ATG 0.7CGG 0.6ACC 0.4AGC 0.3ACT 0.2ACG 0.0.............................................................................................................................................................................SSRs were identi®ed as in Table 14.Figure 30 Duplication l<strong>and</strong>scape <strong>of</strong> chromosome 22. The size <strong>and</strong> location <strong>of</strong>intrachromosomal (blue) <strong>and</strong> interchromosomal (red) duplications are depicted forchromosome 22q, using <strong>the</strong> PARASIGHT computer program (Bailey <strong>and</strong> Eichler,unpublished). Each horizontal line represents 1 Mb (ticks, 100-kb intervals). Thechromosome sequence is oriented from centromere (top left) to telomere (bottom right).Pairwise alignments with . 90% nucleotide identity <strong>and</strong> . 1 kb long are shown. Gapswithin <strong>the</strong> chromosomal sequence are <strong>of</strong> known size <strong>and</strong> shown as empty space.NATURE | VOL 409 | 15 FEBRUARY 2001 | www.nature.com © 2001 Macmillan Magazines Ltd889
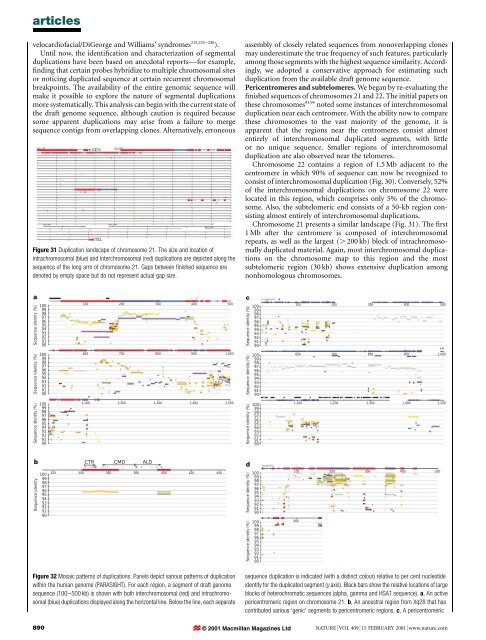
articlesvelocardi<strong>of</strong>acial/DiGeorge <strong>and</strong> Williams' syndromes 215,235±240 ).Until now, <strong>the</strong> identi®cation <strong>and</strong> characterization <strong>of</strong> segmentalduplications have been based on anecdotal reportsÐfor example,®nding that certain probes hybridize to multiple chromosomal sitesor noticing duplicated sequence at certain recurrent chromosomalbreakpoints. The availability <strong>of</strong> <strong>the</strong> entire genomic sequence willmake it possible to explore <strong>the</strong> nature <strong>of</strong> segmental duplicationsmore systematically. This <strong>analysis</strong> can begin with <strong>the</strong> current state <strong>of</strong><strong>the</strong> draft <strong>genome</strong> sequence, although caution is required becausesome apparent duplications may arise from a failure to mergesequence contigs from overlapping clones. Alternatively, erroneousFigure 31 Duplication l<strong>and</strong>scape <strong>of</strong> chromosome 21. The size <strong>and</strong> location <strong>of</strong>intrachromosomal (blue) <strong>and</strong> interchromosomal (red) duplications are depicted along <strong>the</strong>sequence <strong>of</strong> <strong>the</strong> long arm <strong>of</strong> chromosome 21. Gaps between ®nished sequence aredenoted by empty space but do not represent actual gap size.assembly <strong>of</strong> closely related sequences from nonoverlapping clonesmay underestimate <strong>the</strong> true frequency <strong>of</strong> such features, particularlyamong those segments with <strong>the</strong> highest sequence similarity. Accordingly,we adopted a conservative approach for estimating suchduplication from <strong>the</strong> available draft <strong>genome</strong> sequence.Pericentromeres <strong>and</strong> subtelomeres. We began by re-evaluating <strong>the</strong>®nished sequences <strong>of</strong> chromosomes 21 <strong>and</strong> 22. The initial papers on<strong>the</strong>se chromosomes 93,94 noted some instances <strong>of</strong> interchromosomalduplication near each centromere. With <strong>the</strong> ability now to compare<strong>the</strong>se chromosomes to <strong>the</strong> vast majority <strong>of</strong> <strong>the</strong> <strong>genome</strong>, it isapparent that <strong>the</strong> regions near <strong>the</strong> centromeres consist almostentirely <strong>of</strong> interchromosomal duplicated segments, with littleor no unique sequence. Smaller regions <strong>of</strong> interchromosomalduplication are also observed near <strong>the</strong> telomeres.Chromosome 22 contains a region <strong>of</strong> 1.5 Mb adjacent to <strong>the</strong>centromere in which 90% <strong>of</strong> sequence can now be recognized toconsist <strong>of</strong> interchromosomal duplication (Fig. 30). Conversely, 52%<strong>of</strong> <strong>the</strong> interchromosomal duplications on chromosome 22 werelocated in this region, which comprises only 5% <strong>of</strong> <strong>the</strong> chromosome.Also, <strong>the</strong> subtelomeric end consists <strong>of</strong> a 50-kb region consistingalmost entirely <strong>of</strong> interchromosomal duplications.Chromosome 21 presents a similar l<strong>and</strong>scape (Fig. 31). The ®rst1 Mb after <strong>the</strong> centromere is composed <strong>of</strong> interchromosomalrepeats, as well as <strong>the</strong> largest (. 200 kb) block <strong>of</strong> intrachromosomallyduplicated material. Again, most interchromosomal duplicationson <strong>the</strong> chromosome map to this region <strong>and</strong> <strong>the</strong> mostsubtelomeric region (30 kb) shows extensive duplication amongnonhomologous chromosomes.aSequence identity (%)10099989796959493929190100 200 300 400 500cSequence identity (%)10099989796959493929190100 200 300 400 500Sequence identity (%)10099989796959493929190600 700 800 900 1,000Sequence identity (%)10099989796959493929190600 700 800 900 1,000Sequence identity (%)100999897969594939291901,100 1,200 1,300 1,400 1,500Sequence identity (%)100999897969594939291901,100 1,200 1,300 1,400 1,500bSequence identity10099989796959493929190CTR CMD ALD320 340 360 380 420400 440dSequence identity (%)Sequence identity (%)1009998979695949392919010099989796959493929190100 200 300 400 500600Figure 32 Mosaic patterns <strong>of</strong> duplications. Panels depict various patterns <strong>of</strong> duplicationwithin <strong>the</strong> <strong>human</strong> <strong>genome</strong> (PARASIGHT). For each region, a segment <strong>of</strong> draft <strong>genome</strong>sequence (100±500 kb) is shown with both interchromosomal (red) <strong>and</strong> intrachromosomal(blue) duplications displayed along <strong>the</strong> horizontal line. Below <strong>the</strong> line, each separatesequence duplication is indicated (with a distinct colour) relative to per cent nucleotideidentity for <strong>the</strong> duplicated segment (y axis). Black bars show <strong>the</strong> relative locations <strong>of</strong> largeblocks <strong>of</strong> heterochromatic sequences (alpha, gamma <strong>and</strong> HSAT sequence). a, An activepericentromeric region on chromosome 21. b, An ancestral region from Xq28 that hascontributed various `genic' segments to pericentromeric regions. c, A pericentromeric890 © 2001 Macmillan Magazines Ltd NATURE | VOL 409 | 15 FEBRUARY 2001 | www.nature.com