Download pdf - Skog og landskap
Download pdf - Skog og landskap
Download pdf - Skog og landskap
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
as vector for transformation into Norway spruce, it<br />
was termed pCW122*.<br />
pCW122* was re-opened by digestion with Hind III,<br />
the ends were dephosphorylated by CIAP, and PSS1<br />
was ligated into the Hind III sites. The construct was<br />
transformed into E. coli TOP10 cells and ten clones<br />
were obtained. Of these, one proved to have PSS1<br />
inserted in the antisense orientation, the others<br />
contained re-ligated plasmid without insert.<br />
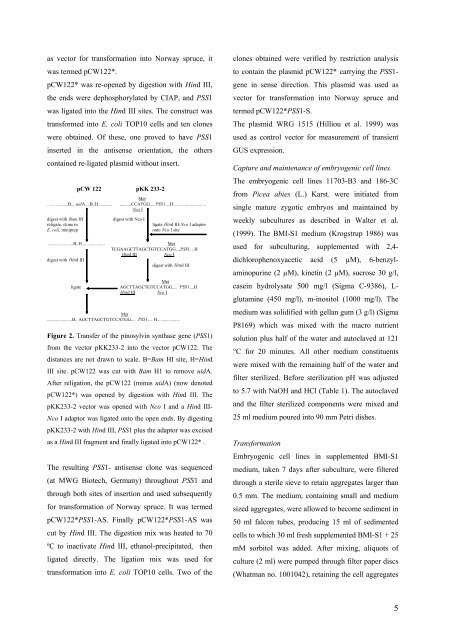
pCW 122<br />
pKK 233-2<br />
Met<br />
………. .......... B.... uid A ....B..H............ ..........CCATGG.... PSS 1 ....H……...... ..............<br />
Nco I<br />
digest with Bam H I digest with Nco I<br />
reli gate, clone in ligate Hin d III - Nco I adaptor<br />
E. coli , miniprep<br />
onto Nco I site<br />
......................B..H....................<br />
digest with Hin d III<br />
ligate<br />
Met<br />
TCGAAGCTTAGCTGTCCATGG....PSS1…H<br />
Hind III Nco I<br />
digest with Hin d III<br />
Met<br />
AGCTTAGCTGTCCATGG.... PSS1....H<br />
Hin d III Nco I<br />
Met<br />
......................B.. AGCTTAGCTGTCCATGG.... PSS 1 .... H....................<br />
Figure 2. Transfer of the pinosylvin synthase gene (PSS1)<br />
from the vector pKK233-2 into the vector pCW122. The<br />
distances are not drawn to scale. B=Bam HI site, H=Hind<br />
III site. pCW122 was cut with Bam H1 to remove uidA.<br />
After religation, the pCW122 (minus uidA) (now denoted<br />
pCW122*) was opened by digestion with Hind III. The<br />
pKK233-2 vector was opened with Nco I and a Hind III-<br />
Nco I adaptor was ligated onto the open ends. By digesting<br />
pKK233-2 with Hind III, PSS1 plus the adaptor was excised<br />
as a Hind III fragment and finally ligated into pCW122* .<br />
The resulting PSS1- antisense clone was sequenced<br />
(at MWG Biotech, Germany) throughout PSS1 and<br />
through both sites of insertion and used subsequently<br />
for transformation of Norway spruce. It was termed<br />
pCW122*PSS1-AS. Finally pCW122*PSS1-AS was<br />
cut by Hind III. The digestion mix was heated to 70<br />
ºC to inactivate Hind III, ethanol-precipitated, then<br />
ligated directly. The ligation mix was used for<br />
transformation into E. coli TOP10 cells. Two of the<br />
clones obtained were verified by restriction analysis<br />
to contain the plasmid pCW122* carrying the PSS1gene<br />
in sense direction. This plasmid was used as<br />
vector for transformation into Norway spruce and<br />
termed pCW122*PSS1-S.<br />
The plasmid WRG 1515 (Hilliou et al. 1999) was<br />
used as control vector for measurement of transient<br />
GUS expression.<br />
Capture and maintenance of embry<strong>og</strong>enic cell lines.<br />
The embry<strong>og</strong>enic cell lines 11703-B3 and 186-3C<br />
from Picea abies (L.) Karst. were initiated from<br />
single mature zygotic embryos and maintained by<br />
weekly subcultures as described in Walter et al.<br />
(1999). The BMI-S1 medium (Kr<strong>og</strong>strup 1986) was<br />
used for subculturing, supplemented with 2,4dichlorophenoxyacetic<br />
acid (5 µM), 6-benzylaminopurine<br />
(2 µM), kinetin (2 µM), sucrose 30 g/l,<br />
casein hydrolysate 500 mg/l (Sigma C-9386), Lglutamine<br />
(450 mg/l), m-inositol (1000 mg/l). The<br />
medium was solidified with gellan gum (3 g/l) (Sigma<br />
P8169) which was mixed with the macro nutrient<br />
solution plus half of the water and autoclaved at 121<br />
°C for 20 minutes. All other medium constituents<br />
were mixed with the remaining half of the water and<br />
filter sterilized. Before sterilization pH was adjusted<br />
to 5.7 with NaOH and HCl (Table 1). The autoclaved<br />
and the filter sterilized components were mixed and<br />
25 ml medium poured into 90 mm Petri dishes.<br />
Transformation<br />
Embry<strong>og</strong>enic cell lines in supplemented BMI-S1<br />
medium, taken 7 days after subculture, were filtered<br />
through a sterile sieve to retain aggregates larger than<br />
0.5 mm. The medium, containing small and medium<br />
sized aggregates, were allowed to become sediment in<br />
50 ml falcon tubes, producing 15 ml of sedimented<br />
cells to which 30 ml fresh supplemented BMI-S1 + 25<br />
mM sorbitol was added. After mixing, aliquots of<br />
culture (2 ml) were pumped through filter paper discs<br />
(Whatman no. 1001042), retaining the cell aggregates<br />
5