Peptidoglycan .Types of Bacterial Cell Walls and their Taxonomic ...
Peptidoglycan .Types of Bacterial Cell Walls and their Taxonomic ...
Peptidoglycan .Types of Bacterial Cell Walls and their Taxonomic ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
VOL. 36, 1972<br />
PEPTIDOGLYCAN TYPES OF BACTERIAL CELL WALLS<br />
first discovered by Strange <strong>and</strong> Dark (364).<br />
Further studies have shown that it is the<br />
3-O-D-lactic acid ether <strong>of</strong> glucosamine (365,<br />
384) (Fig. 1). The glycan reveals only few<br />
variations, such as acetylation or phosphorylation<br />
<strong>of</strong> the muramyl 6-hydroxyl groups (9, 109,<br />
224) <strong>and</strong> the occasional absence <strong>of</strong> a peptide or<br />
N-acetyl substituent (10, 109). Recent studies<br />
on the peptidoglycan <strong>of</strong> bacterial spores have<br />
indicated that muramic acid residues can be<br />
present in the muramic lactam form, a sugar<br />
not previously found in nature <strong>and</strong>, hence, a<br />
unique spore constituent (401). Among mycobacteria<br />
(3, 25), Nocardia kirovani (130) <strong>and</strong><br />
Micromonospora (398), muramic acid does not<br />
occur as N-acetyl, but as the N-glycolyl derivative.<br />
Here the amino group in position 2 is not<br />
substituted by an acetyl group (-COCH3) but<br />
by a glycolyl group (-COCH2OH). Studies on<br />
a wide variety <strong>of</strong> gram-positive <strong>and</strong> gram-negative<br />
bacteria indicated that only glucomuramic<br />
acid occurs in the wall: galactomuramic acid<br />
has not been found so far (413, 414).<br />
The chain length <strong>of</strong> the glycans has been<br />
discussed in detail by Ghuysen (109). The<br />
glycans are polydisperse <strong>and</strong> thus only average<br />
figures can be given. In different organisms the<br />
average chain length varies between 10 <strong>and</strong> 65<br />
disaccharide units (109, 148). Although there is<br />
a relationship between the cell shape <strong>and</strong> average<br />
chain length <strong>of</strong> the glycan in some special<br />
cases (210), there is no evidence for a general<br />
correlation (204, 402).<br />
Peptide moiety. The peptide moiety is<br />
bound through its N terminus to the carboxyl<br />
group <strong>of</strong> muramic acid <strong>and</strong> contains alternating<br />
L <strong>and</strong> n amino acids. The occurrence <strong>of</strong> amino<br />
acids with the D configuration is a typical<br />
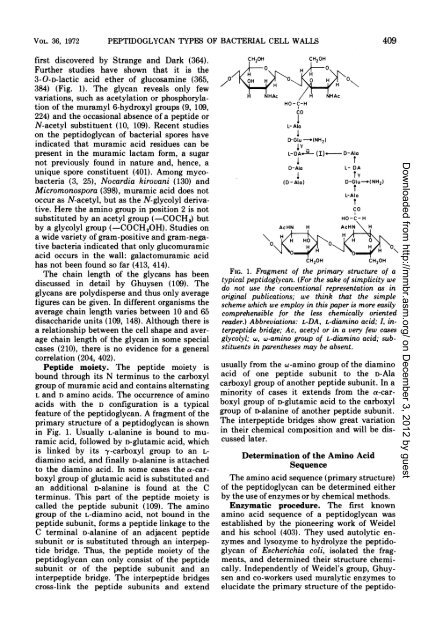
feature <strong>of</strong> the peptidoglycan. A fragment <strong>of</strong> the<br />
primary structure <strong>of</strong> a peptidoglycan is shown<br />
in Fig. 1. Usually L-alanine is bound to muramic<br />
acid, followed by r)-glutamic acid, which<br />
is linked by its y-carboxyl group to an Ldiamino<br />
acid, <strong>and</strong> finally D-alanine is attached<br />
to the diamino acid. In some cases the a-carboxyl<br />
group <strong>of</strong> glutamic acid is substituted <strong>and</strong><br />
an additional D-alanine is found at the C<br />
terminus. This part <strong>of</strong> the peptide moiety is<br />
called the peptide subunit (109). The amino<br />
group <strong>of</strong> the L-diamino acid, not bound in the<br />
peptide subunit, forms a peptide linkage to the<br />
C terminal D-alanine <strong>of</strong> an adjacent peptide<br />
subunit or is substituted through an interpeptide<br />
bridge. Thus, the peptide moiety <strong>of</strong> the<br />
peptidoglycan can only consist <strong>of</strong> the peptide<br />
subunit or <strong>of</strong> the peptide subunit <strong>and</strong> an<br />
interpeptide bridge. The interpeptide bridges<br />
cross-link the peptide subunits <strong>and</strong> extend<br />
CH20H CH20H<br />
0 H<br />
OH H 0 0 H 0<br />
HAc H HAc<br />
HO-C-H<br />
CO<br />
I<br />
L- Ala<br />
4<br />
D-Glu -(NH2)<br />
4Y -<br />
L-tDA ---- (I)- D-Ala<br />
4 t<br />
D-Ala L- DA<br />
4 Y<br />
(D-Ala) D-Glu-(NH2)<br />
L-Ala<br />
409<br />
CO<br />
HO-C- H<br />
AcHN H AcHN H<br />
H HO H<br />
~~H0 H 0 H<br />
H H 0<br />
CH20H CH20H<br />
FIG. 1. Fragment <strong>of</strong> the primary structure <strong>of</strong> a<br />
typical peptidoglycan. (For the sake <strong>of</strong> simplicity we<br />
do not use the conventional representation as in<br />
original publications; we think that the simple<br />
scheme which we employ in this paper is more easily<br />
comprehensible for the less chemically oriented<br />
reader.) Abbreviations: L-DA, L-diamino acid; I, interpeptide<br />
bridge; Ac, acetyl or in a very few cases<br />
glycolyl; w, w-amino group <strong>of</strong> L-diamino acid; substituents<br />
in parentheses may be absent.<br />
usually from the w-amino group <strong>of</strong> the diamino<br />
acid <strong>of</strong> one peptide subunit to the D-Ala<br />
carboxyl group <strong>of</strong> another peptide subunit. In a<br />
minority <strong>of</strong> cases it extends from the a-carboxyl<br />
group <strong>of</strong> D-glutamic acid to the carboxyl<br />
group <strong>of</strong> -alanine <strong>of</strong> another peptide subunit.<br />
The interpeptide bridges show great variation<br />
in <strong>their</strong> chemical composition <strong>and</strong> will be discussed<br />
later.<br />
Determination <strong>of</strong> the Amino Acid<br />
Sequence<br />
The amino acid sequence (primary structure)<br />
<strong>of</strong> the peptidoglycan can be determined either<br />
by the use <strong>of</strong> enzymes or by chemical methods.<br />
Enzymatic procedure. The first known<br />
amino acid sequence <strong>of</strong> a peptidoglycan was<br />
established by the pioneering work <strong>of</strong> Weidel<br />
<strong>and</strong> his school (403). They used autolytic enzymes<br />
<strong>and</strong> lysozyme to hydrolyze the peptidoglycan<br />
<strong>of</strong> Escherichia coli, isolated the fragments,<br />
<strong>and</strong> determined <strong>their</strong> structure chemically.<br />
Independently <strong>of</strong> Weidel's group, Ghuysen<br />
<strong>and</strong> co-workers used muralytic enzymes to<br />
elucidate the primary structure <strong>of</strong> the peptido-<br />
Downloaded from<br />
http://mmbr.asm.org/<br />
on December 3, 2012 by guest