TGQR 2010Q2 Report.pdf - Teragridforum.org
TGQR 2010Q2 Report.pdf - Teragridforum.org
TGQR 2010Q2 Report.pdf - Teragridforum.org
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
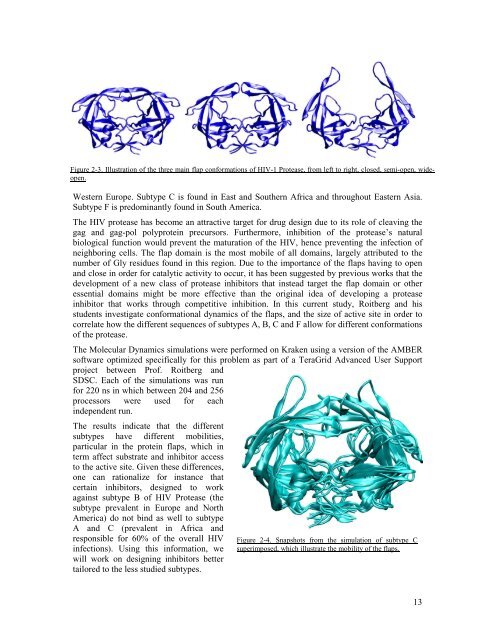
Figure 2-3. Illustration of the three main flap conformations of HIV-1 Protease, from left to right, closed, semi-open, wideopen.<br />
Western Europe. Subtype C is found in East and Southern Africa and throughout Eastern Asia.<br />
Subtype F is predominantly found in South America.<br />
The HIV protease has become an attractive target for drug design due to its role of cleaving the<br />
gag and gag-pol polyprotein precursors. Furthermore, inhibition of the protease’s natural<br />
biological function would prevent the maturation of the HIV, hence preventing the infection of<br />
neighboring cells. The flap domain is the most mobile of all domains, largely attributed to the<br />
number of Gly residues found in this region. Due to the importance of the flaps having to open<br />
and close in order for catalytic activity to occur, it has been suggested by previous works that the<br />
development of a new class of protease inhibitors that instead target the flap domain or other<br />
essential domains might be more effective than the original idea of developing a protease<br />
inhibitor that works through competitive inhibition. In this current study, Roitberg and his<br />
students investigate conformational dynamics of the flaps, and the size of active site in order to<br />
correlate how the different sequences of subtypes A, B, C and F allow for different conformations<br />
of the protease.<br />
The Molecular Dynamics simulations were performed on Kraken using a version of the AMBER<br />
software optimized specifically for this problem as part of a TeraGrid Advanced User Support<br />
project between Prof. Roitberg and<br />
SDSC. Each of the simulations was run<br />
for 220 ns in which between 204 and 256<br />
processors were used for each<br />
independent run.<br />
The results indicate that the different<br />
subtypes have different mobilities,<br />
particular in the protein flaps, which in<br />
term affect substrate and inhibitor access<br />
to the active site. Given these differences,<br />
one can rationalize for instance that<br />
certain inhibitors, designed to work<br />
against subtype B of HIV Protease (the<br />
subtype prevalent in Europe and North<br />
America) do not bind as well to subtype<br />
A and C (prevalent in Africa and<br />
responsible for 60% of the overall HIV<br />
infections). Using this information, we<br />
will work on designing inhibitors better<br />
tailored to the less studied subtypes.<br />
Figure 2-4. Snapshots from the simulation of subtype C<br />
superimposed, which illustrate the mobility of the flaps.<br />
13