Biennial Report 2011â2012
Biennial Report 2011â2012
Biennial Report 2011â2012
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Department of Protein-Nucleic Acids Interactions<br />
switches its orientation on DNA to match the polarity of the<br />
second strand and then cuts the phosphodiester bond on the<br />
second DNA strand. Surprisingly, we find that an enzyme flip<br />
required for the second DNA strand cleavage occurs without<br />
an excursion into bulk solution, as the same BcnI molecule acts<br />
processively on both DNA strands. We provide evidence that<br />
after cleavage of the first DNA strand, BcnI remains associated<br />
with the nicked intermediate and relocates to the opposite<br />
strand by a short range diffusive hopping on DNA.<br />
Structure and molecular mechanisms of<br />
CRISPR/Cas systems: projects overview<br />
Streptococcus thermophilus DGCC7710 strain, for which biological<br />
activity of the CRISPR/Cas system has been directly<br />
demonstrated in a phage challenge assay, contains four distinct<br />
systems: CRISPR1, CRISPR2, CRISPR3 and CRISPR4,<br />
which belong to the three distinct Types. Direct spacer incorporation<br />
activity has been demonstrated for the CRISPR1 and<br />
CRISPR3 systems, with the former being more active. The<br />
CRISPR2 system seems to be disrupted and non-functional,<br />
whilst functional activity of CRISPR4 has not yet been demonstrated.<br />
Cas genes, which are specific to the repeat regions<br />
and fall into different families, are located in close proximity to<br />
the spacer-repeat region and encode proteins that often carry<br />
functional nucleic-acid related domains such as nucleases, helicases,<br />
polymerases and nucleotide binding. We aim to characterize<br />
the functional and biochemical activities of Cas proteins<br />
belonging to the CRISPR1, CRISPR3 and CRISPR4 systems<br />
of S. thermophilus.<br />
RNA-guided DNA endonuclease provides<br />
DNA silencing in the Type II system<br />
Type II CRISPR-Cas systems typically consist of only four<br />
Cas genes. The mechanism for DNA interference provided<br />
by the Type II systems remained to be established. We show<br />
that in the CRISPR3 system of Streptococcus thermophilus (a<br />
model and active Type II CRISPR/Cas system), Cas9 associates<br />
with crRNA to form an effector complex which specifically<br />
cleaves matching target dsDNA. This contrasts sharply<br />
with effector complexes for Type I and Type III systems,<br />
which are multisubunit ribonucleoprotein complexes. We<br />
isolated the Cas9-crRNA complex and demonstrated that it<br />
generates in vitro a double strand break at specific sites in<br />
target DNA molecules that are complementary to crRNA sequences<br />
and bear a short proto-spacer adjacent motif (PAM),<br />
in the direct vicinity of the matching sequence. We show that<br />
DNA cleavage is executed by two distinct active sites (RuvC<br />
and HNH) within Cas9, to generate site-specific nicks on opposite<br />
DNA strands. Sequence specificity of the Cas9-crRNA<br />
complex is dictated by the 42 nt crRNA which includes a<br />
20 nt fragment complementary to the proto-spacer sequence<br />
in the target DNA. All together our data demonstrate that<br />
the Cas9-crRNA complex functions as an RNA-guided endonuclease<br />
with sequence-specific target site recognition and<br />
cleavage through two distinct strand nicks.<br />
Figure 5. CRISPR/Cas systems of S. thermophilus DGCC7710.<br />
CRISPR1 and CRISPR3 systems belong to the TypeII, CRISPR2 to the<br />
TypeIII whilst CRISPR4 belongs to the Type I (E.coli subtype).<br />
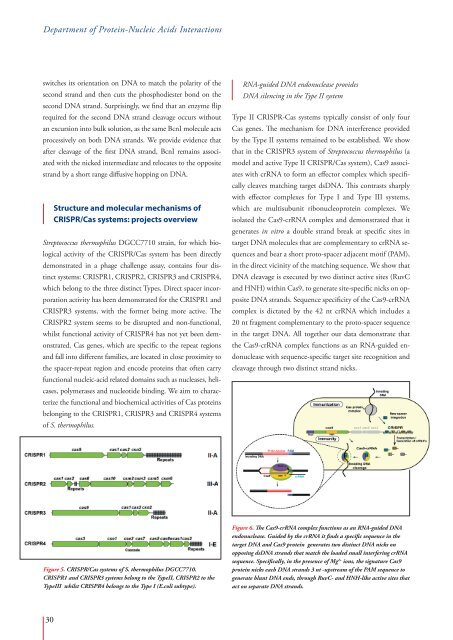
Figure 6. The Cas9-crRNA complex functions as an RNA-guided DNA<br />
endonuclease. Guided by the crRNA it finds a specific sequence in the<br />
target DNA and Cas9 protein generates two distinct DNA nicks on<br />
opposing dsDNA strands that match the loaded small interfering crRNA<br />
sequence. Specifically, in the presence of Mg 2+ ions, the signature Cas9<br />
protein nicks each DNA strands 3 nt -upstream of the PAM sequence to<br />
generate blunt DNA ends, through RuvC- and HNH-like active sites that<br />
act on separate DNA strands.<br />
30