Biennial Report 2011â2012
Biennial Report 2011â2012
Biennial Report 2011â2012
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Engineering the catalytic reaction<br />
of methyltransferases for targeted<br />
covalent labeling of DNA<br />
G. Lukinavičius, V. Masevičius,<br />
G. Urbanavičiūtė, R. Gerasimaitė, M. Tomkuvienė<br />
We had synthesized a series of model AdoMet analogs with<br />
sulfonium-bound extended side chains replacing the methyl<br />
group and showed that allylic and propargylic side chains<br />
can be efficiently transferred by DNA MTases with high sequence-<br />
and base-specificity (Dalhoff et al, Nature Chem.<br />
Biol., 2006, 2: 31–32; Klimašauskas and Weinhold, Trends<br />
Biotechnol., 2007, 25: 99–104) in collaboration with the<br />
group of Prof. Elmar Weinhold (RWTH Aachen, Germany).<br />
Using DNA MTases along with their novel cofactors that carry<br />
useful functional or reporter groups, we demonstrated that<br />
our new approach name mTAG (methyltransferase-directed<br />
Transfer of Activated Groups) can be used for sequence-specific<br />
functionalization and labeling of a wide variety of model<br />
and natural DNA substrates (Lukinavicius et al., J. Amer.<br />
Chem. Soc., 2007, 129: 2758).<br />
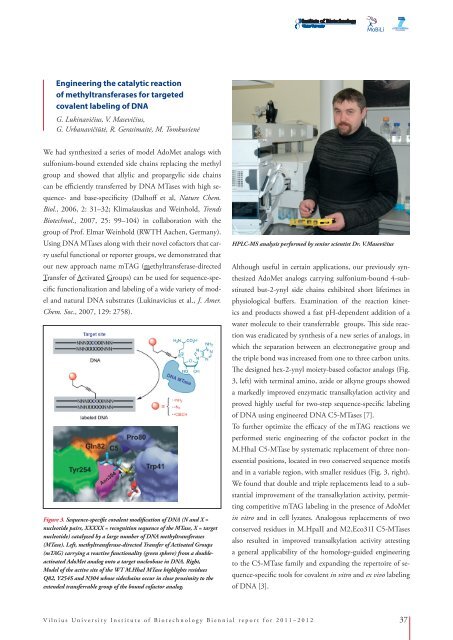
Figure 3. Sequence-specific covalent modification of DNA (N and X =<br />
nucleotide pairs, XXXXX = recognition sequence of the MTase, X = target<br />
nucleotide) catalyzed by a large number of DNA methyltransferases<br />
(MTase). Left, methyltransferase-directed Transfer of Activated Groups<br />
(mTAG) carrying a reactive functionality (green sphere) from a doubleactivated<br />
AdoMet analog onto a target nucleobase in DNA. Right,<br />
Model of the active site of the WT M.HhaI MTase highlights residues<br />
Q82, Y254S and N304 whose sidechains occur in close proximity to the<br />
extended transferrable group of the bound cofactor analog.<br />
HPLC-MS analysis performed by senior scientist Dr. V.Masevičius<br />
Although useful in certain applications, our previously synthesized<br />
AdoMet analogs carrying sulfonium-bound 4-substituted<br />
but-2-ynyl side chains exhibited short lifetimes in<br />
physiological buffers. Examination of the reaction kinetics<br />
and products showed a fast pH-dependent addition of a<br />
water molecule to their transferrable groups. This side reaction<br />
was eradicated by synthesis of a new series of analogs, in<br />
which the separation between an electronegative group and<br />
the triple bond was increased from one to three carbon units.<br />
The designed hex-2-ynyl moiety-based cofactor analogs (Fig.<br />
3, left) with terminal amino, azide or alkyne groups showed<br />
a markedly improved enzymatic transalkylation activity and<br />
proved highly useful for two-step sequence-specific labeling<br />
of DNA using engineered DNA C5-MTases [7].<br />
To further optimize the efficacy of the mTAG reactions we<br />
performed steric engineering of the cofactor pocket in the<br />
M.HhaI C5-MTase by systematic replacement of three nonessential<br />
positions, located in two conserved sequence motifs<br />
and in a variable region, with smaller residues (Fig. 3, right).<br />
We found that double and triple replacements lead to a substantial<br />
improvement of the transalkylation activity, permitting<br />
competitive mTAG labeling in the presence of AdoMet<br />
in vitro and in cell lyzates. Analogous replacements of two<br />
conserved residues in M.HpaII and M2.Eco31I C5-MTases<br />
also resulted in improved transalkylation activity attesting<br />
a general applicability of the homology-guided engineering<br />
to the C5-MTase family and expanding the repertoire of sequence-specific<br />
tools for covalent in vitro and ex vivo labeling<br />
of DNA [3].<br />
Vilnius University Institute of Biotechnology <strong>Biennial</strong> report for 2011–2012 37