Biennial Report 2011â2012
Biennial Report 2011â2012
Biennial Report 2011â2012
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
At present computational methods are playing an increasingly<br />
important role in biological research. Breakthroughs in<br />
technologies have resulted in a flood of various types of biological<br />
data such as genome sequences for different organisms,<br />
data on gene expression, protein-protein interactions,<br />
etc. Computational biology and bioinformatics are helping to<br />
make sense of all this vast biological data by providing tools<br />
for performing large-scale studies. In addition, computational<br />
biologists are utilizing available experimental data to improve<br />
various analytical and predictive methods that could help address<br />
specific biological problems.<br />
Research carried out in our department covers a broad range of<br />
topics that together can be described as Computational Studies<br />
of Protein Structure, Function and Evolution. There are two main<br />
research directions:<br />
Development of methods for the detection of protein homology<br />
from sequence data, comparative modeling, analysis<br />
and evaluation of protein three-dimensional structure.<br />
Application of computational methods for discovering<br />
general patterns in biological data, structural/functional characterization<br />
of proteins and their complexes; design of novel<br />
proteins and mutants with desired properties. We address a variety<br />
of challenging biological problems, yet our main focus is<br />
on proteins and protein complexes involved in DNA replication,<br />
repair and recombination.<br />
Development of<br />
computational methods<br />
At present, we are in the final stages of the development of a<br />
new competitive homology detection method.<br />
The evaluation of protein structure is particularly important in<br />
computational protein modeling. Scoring models against the<br />
native structure is at the heart of development and benchmarking<br />
of protein structure prediction and refinement methods. It<br />
may seem that one-to-one correspondence between computational<br />
models and the native (reference) structure should make<br />
such evaluation trivial. Yet, contrary to this view, it is an open<br />
problem, because many aspects of the reference-based model<br />
evaluation still lack desired robustness. We have been actively<br />
researching how to improve the reference-based model evaluation.<br />
Our attempts resulted in a new highly effective score,<br />
which is described in more detail below.<br />
CAD-score: evaluation of protein structural<br />
models based on contact area difference<br />
CAD-score (Contact Area Difference score) is a new evaluation<br />
function quantifying differences between physical contacts<br />
in a model and the reference structure. It uses the concept<br />
of residue-residue contact area difference (CAD) introduced<br />
by Abagyan & Totrov (J. Mol. Biol. 1997; 268:678–<br />
685). Contact areas, the underlying basis of the score, are derived<br />
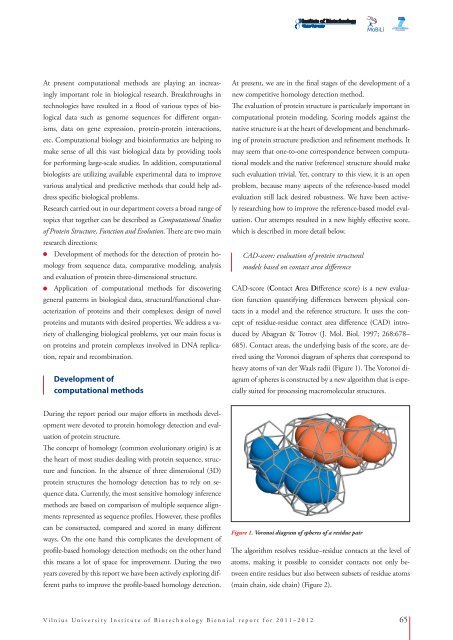
using the Voronoi diagram of spheres that correspond to<br />
heavy atoms of van der Waals radii (Figure 1). The Voronoi diagram<br />
of spheres is constructed by a new algorithm that is especially<br />
suited for processing macromolecular structures.<br />
During the report period our major efforts in methods development<br />
were devoted to protein homology detection and evaluation<br />
of protein structure.<br />
The concept of homology (common evolutionary origin) is at<br />
the heart of most studies dealing with protein sequence, structure<br />
and function. In the absence of three dimensional (3D)<br />
protein structures the homology detection has to rely on sequence<br />
data. Currently, the most sensitive homology inference<br />
methods are based on comparison of multiple sequence alignments<br />
represented as sequence profiles. However, these profiles<br />
can be constructed, compared and scored in many different<br />
ways. On the one hand this complicates the development of<br />
profile-based homology detection methods; on the other hand<br />
this means a lot of space for improvement. During the two<br />
years covered by this report we have been actively exploring different<br />
paths to improve the profile-based homology detection.<br />
Figure 1. Voronoi diagram of spheres of a residue pair<br />
The algorithm resolves residue–residue contacts at the level of<br />
atoms, making it possible to consider contacts not only between<br />
entire residues but also between subsets of residue atoms<br />
(main chain, side chain) (Figure 2).<br />
Vilnius University Institute of Biotechnology <strong>Biennial</strong> report for 2011–2012 65