Ensembl Compara - CNB - Protein Design Group
Ensembl Compara - CNB - Protein Design Group
Ensembl Compara - CNB - Protein Design Group
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
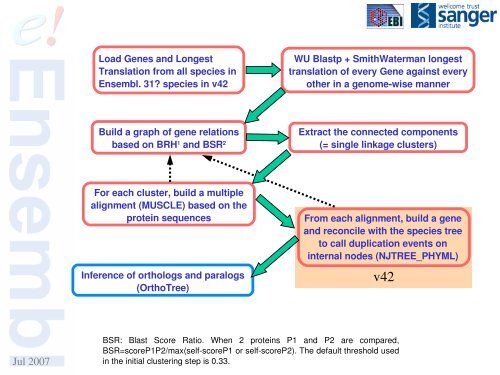
Load Genes and LongestTranslation from all species in<strong>Ensembl</strong>. 31? species in v42WU Blastp + SmithWaterman longesttranslation of every Gene against everyother in a genomewise mannerBuild a graph of gene relationsbased on BRH 1 and BSR 2Extract the connected components(= single linkage clusters)For each cluster, build a multiplealignment (MUSCLE) based on theprotein sequencesInference of orthologs and paralogs(OrthoTree)From each alignment, build a geneand reconcile with the species treeto call duplication events oninternal nodes (NJTREE_PHYML)v42Jul 2007BSR: Blast Score Ratio. When 2 proteins P1 and P2 are compared,BSR=scoreP1P2/max(selfscoreP1 or selfscoreP2). The default threshold usedin the initial clustering step is 0.33.