Etudes sur le mécanisme de remodelage des nucléosomes par ...
Etudes sur le mécanisme de remodelage des nucléosomes par ...
Etudes sur le mécanisme de remodelage des nucléosomes par ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
tel-00413908, version 1 - 7 Sep 2009<br />
HaeIII<br />
time<br />
A B<br />
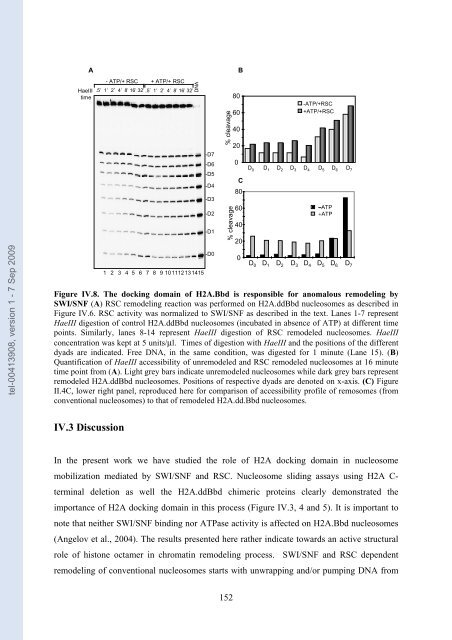
Figure IV.8. The docking domain of H2A.Bbd is responsib<strong>le</strong> for anomalous remo<strong>de</strong>ling by<br />
SWI/SNF (A) RSC remo<strong>de</strong>ling reaction was performed on H2A.ddBbd nuc<strong>le</strong>osomes as <strong>de</strong>scribed in<br />
Figure IV.6. RSC activity was normalized to SWI/SNF as <strong>de</strong>scribed in the text. Lanes 1-7 represent<br />
HaeIII digestion of control H2A.ddBbd nuc<strong>le</strong>osomes (incubated in absence of ATP) at different time<br />
points. Similarly, lanes 8-14 represent HaeIII digestion of RSC remo<strong>de</strong><strong>le</strong>d nuc<strong>le</strong>osomes. HaeIII<br />
concentration was kept at 5 units/μl. Times of digestion with HaeIII and the positions of the different<br />
dyads are indicated. Free DNA, in the same condition, was digested for 1 minute (Lane 15). (B)<br />
Quantification of HaeIII accessibility of unremo<strong>de</strong><strong>le</strong>d and RSC remo<strong>de</strong><strong>le</strong>d nuc<strong>le</strong>osomes at 16 minute<br />
time point from (A). Light grey bars indicate unremo<strong>de</strong><strong>le</strong>d nuc<strong>le</strong>osomes whi<strong>le</strong> dark grey bars represent<br />
remo<strong>de</strong><strong>le</strong>d H2A.ddBbd nuc<strong>le</strong>osomes. Positions of respective dyads are <strong>de</strong>noted on x-axis. (C) Figure<br />
II.4C, lower right panel, reproduced here for com<strong>par</strong>ison of accessibility profi<strong>le</strong> of remosomes (from<br />
conventional nuc<strong>le</strong>osomes) to that of remo<strong>de</strong><strong>le</strong>d H2A.dd.Bbd nuc<strong>le</strong>osomes.<br />
IV.3 Discussion<br />
- ATP/+ RSC + ATP/+ RSC<br />
.5’ 1’ 2’ 4’ 8’ 16’ 32’ .5’ 1’ 2’ 4’ 8’ 16’ 32’ DNA<br />
1 2 3 4 5 6 7 8 9 10 1112 13 1415<br />
-D7<br />
-D6<br />
-D5<br />
-D4<br />
-D3<br />
-D2<br />
-D1<br />
-D0<br />
% c<strong>le</strong>avage<br />
% c<strong>le</strong>avage<br />
In the present work we have studied the ro<strong>le</strong> of H2A docking domain in nuc<strong>le</strong>osome<br />
mobilization mediated by SWI/SNF and RSC. Nuc<strong>le</strong>osome sliding assays using H2A C-<br />
terminal <strong>de</strong><strong>le</strong>tion as well the H2A.ddBbd chimeric proteins c<strong>le</strong>arly <strong>de</strong>monstrated the<br />
importance of H2A docking domain in this process (Figure IV.3, 4 and 5). It is important to<br />
note that neither SWI/SNF binding nor ATPase activity is affected on H2A.Bbd nuc<strong>le</strong>osomes<br />
(Angelov et al., 2004). The results presented here rather indicate towards an active structural<br />
ro<strong>le</strong> of histone octamer in chromatin remo<strong>de</strong>ling process. SWI/SNF and RSC <strong>de</strong>pen<strong>de</strong>nt<br />
remo<strong>de</strong>ling of conventional nuc<strong>le</strong>osomes starts with unwrapping and/or pumping DNA from<br />
152<br />
80<br />
60<br />
40<br />
20<br />
0<br />
C<br />
80<br />
60<br />
40<br />
20<br />
0<br />
-ATP/+RSC<br />
+ATP/+RSC<br />
D 0 D 1 D 2 D 3 D 4 D 5 D 6 D 7<br />
D 0<br />
D 1<br />
D 2<br />
D 3<br />
D 4<br />
−ATP<br />
+ATP<br />
D 5<br />
D 6<br />
D 7