- Page 4:
GENE CLONING AND DNA ANALYSIS
- Page 10:
This edition first published 2010,
- Page 16:
Contents CONTENTS Preface to the Si
- Page 20:
Contents ix 4.3 Ligation—joining
- Page 24:
Contents xi 8 How to Obtain a Clone
- Page 28:
Contents xiii 11.3 Identifying and
- Page 32:
Contents xv 15.1.2 Herbicide resist
- Page 36:
PART I The Basic Principles of Gene
- Page 42:
4 Part I The Basic Principles of Ge
- Page 46:
6 1.4 What is PCR? Figure 1.2 The b
- Page 50:
8 Figure 1.3 Cloning allows individ
- Page 54:
10 Figure 1.5 Gene isolation by PCR
- Page 58:
12 Further reading FURTHER READING
- Page 62:
14 Figure 2.1 Plasmids: independent
- Page 66:
16 Figure 2.4 Plasmid transfer by c
- Page 70:
18 Figure 2.6 Capsid components Pha
- Page 74:
20 Figure 2.7 λ phage particle att
- Page 78:
22 (a) The linear form of the λ DN
- Page 82:
24 Part I The Basic Principles of G
- Page 86:
26 Bacterial culture Table 3.1 Cent
- Page 90:
28 Bacterial culture Figure 3.3 Har
- Page 94:
30 Figure 3.6 Removal of protein co
- Page 98:
32 Figure 3.8 Collecting DNA by eth
- Page 102: 34 Figure 3.10 DNA purification by
- Page 106: 36 Figure 3.12 Two conformations of
- Page 110: 38 Figure 3.15 Partial unwinding of
- Page 114: 40 Figure 3.18 Preparation of a pha
- Page 118: 42 Figure 3.21 Collection of phage
- Page 122: 44 Further reading FURTHER READING
- Page 126: 46 Part I The Basic Principles of G
- Page 130: 48 Figure 4.3 The reactions catalyz
- Page 134: 50 Figure 4.6 (a) Alkaline phosphat
- Page 138: 52 Figure 4.8 (a) Restriction of ph
- Page 142: 54 Figure 4.9 (a) Production of blu
- Page 146: 56 Table 4.2 A 10 × buffer suitabl
- Page 150: 58 Figure 4.13 Visualizing DNA band
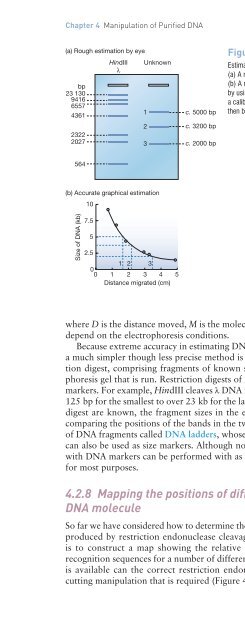
- Page 156: Chapter 4 Manipulation of Purified
- Page 160: Chapter 4 Manipulation of Purified
- Page 164: Chapter 4 Manipulation of Purified
- Page 168: Chapter 4 Manipulation of Purified
- Page 172: Chapter 4 Manipulation of Purified
- Page 176: Chapter 4 Manipulation of Purified
- Page 180: Chapter 5 Introduction of DNA into
- Page 184: Chapter 5 Introduction of DNA into
- Page 188: Chapter 5 Introduction of DNA into
- Page 192: Chapter 5 Introduction of DNA into
- Page 196: Chapter 5 Introduction of DNA into
- Page 200: Chapter 5 Introduction of DNA into
- Page 204:
Chapter 5 Introduction of DNA into
- Page 208:
Chapter 5 Introduction of DNA into
- Page 212:
Chapter 6 Cloning Vectors for E. co
- Page 216:
Chapter 6 Cloning Vectors for E. co
- Page 220:
Chapter 6 Cloning Vectors for E. co
- Page 224:
Chapter 6 Cloning Vectors for E. co
- Page 228:
Chapter 6 Cloning Vectors for E. co
- Page 232:
Chapter 6 Cloning Vectors for E. co
- Page 236:
Chapter 6 Cloning Vectors for E. co
- Page 240:
Chapter 6 Cloning Vectors for E. co
- Page 244:
Chapter 7 Cloning Vectors for Eukar
- Page 248:
Chapter 7 Cloning Vectors for Eukar
- Page 252:
Chapter 7 Cloning Vectors for Eukar
- Page 256:
Chapter 7 Cloning Vectors for Eukar
- Page 260:
Chapter 7 Cloning Vectors for Eukar
- Page 264:
Chapter 7 Cloning Vectors for Eukar
- Page 268:
Chapter 7 Cloning Vectors for Eukar
- Page 272:
Chapter 7 Cloning Vectors for Eukar
- Page 276:
Chapter 7 Cloning Vectors for Eukar
- Page 280:
Chapter 7 Cloning Vectors for Eukar
- Page 284:
Chapter 7 Cloning Vectors for Eukar
- Page 288:
Chapter 8 How to Obtain a Clone of
- Page 292:
Chapter 8 How to Obtain a Clone of
- Page 296:
Chapter 8 How to Obtain a Clone of
- Page 300:
Chapter 8 How to Obtain a Clone of
- Page 304:
Chapter 8 How to Obtain a Clone of
- Page 308:
Chapter 8 How to Obtain a Clone of
- Page 312:
Chapter 8 How to Obtain a Clone of
- Page 316:
Chapter 8 How to Obtain a Clone of
- Page 320:
Chapter 8 How to Obtain a Clone of
- Page 324:
Chapter 8 How to Obtain a Clone of
- Page 328:
Chapter 9 The Polymerase Chain Reac
- Page 332:
Chapter 10 The Polymerase Chain Rea
- Page 336:
Chapter 10 The Polymerase Chain Rea
- Page 340:
Chapter 10 The Polymerase Chain Rea
- Page 344:
Chapter 10 The Polymerase Chain Rea
- Page 348:
Chapter 10 The Polymerase Chain Rea
- Page 352:
Chapter 10 The Polymerase Chain Rea
- Page 356:
Chapter 10 The Polymerase Chain Rea
- Page 364:
Chapter 10 Sequencing Genes and Gen
- Page 368:
Chapter 10 Sequencing Genes and Gen
- Page 372:
Chapter 10 Sequencing Genes and Gen
- Page 376:
Chapter 10 Sequencing Genes and Gen
- Page 380:
Chapter 10 Sequencing Genes and Gen
- Page 384:
Chapter 10 Sequencing Genes and Gen
- Page 388:
Chapter 10 Sequencing Genes and Gen
- Page 392:
Chapter 10 Sequencing Genes and Gen
- Page 396:
Chapter 10 Sequencing Genes and Gen
- Page 400:
Chapter 10 Sequencing Genes and Gen
- Page 404:
Chapter 11 Studying Gene Expression
- Page 408:
Chapter 11 Studying Gene Expression
- Page 412:
Chapter 11 Studying Gene Expression
- Page 416:
Chapter 11 Studying Gene Expression
- Page 420:
Chapter 11 Studying Gene Expression
- Page 424:
Chapter 11 Studying Gene Expression
- Page 428:
Chapter 11 Studying Gene Expression
- Page 432:
Chapter 11 Studying Gene Expression
- Page 436:
Chapter 11 Studying Gene Expression
- Page 440:
Chapter 11 Studying Gene Expression
- Page 444:
Chapter 11 Studying Gene Expression
- Page 448:
Chapter 12 Studying Genomes Chapter
- Page 452:
Chapter 12 Studying Genomes 209 Fig
- Page 456:
Chapter 12 Studying Genomes 211 Fig
- Page 460:
Chapter 12 Studying Genomes 213 (a)
- Page 464:
Chapter 12 Studying Genomes 215 12.
- Page 468:
Chapter 12 Studying Genomes 217 C G
- Page 472:
Chapter 12 Studying Genomes 219 Fig
- Page 476:
Chapter 12 Studying Genomes 221 Fig
- Page 480:
PART III The Applications of Gene C
- Page 486:
226 Figure 13.1 Two different syste
- Page 490:
228 Figure 13.3 The three most impo
- Page 494:
230 Figure 13.6 Strong and weak pro
- Page 498:
232 Part III The Applications of Ge
- Page 502:
234 Figure 13.12 Fusion protein bou
- Page 506:
236 organism has a bias toward pref
- Page 510:
238 Figure 13.16 Part III The Appli
- Page 514:
240 Figure 13.18 Crystalline inclus
- Page 518:
242 Figure 13.20 Recombinant protei
- Page 522:
244 s Part III The Applications of
- Page 526:
246 14.1.1 Recombinant insulin Figu
- Page 530:
248 Figure 14.2 The synthesis of re
- Page 534:
250 Figure 14.5 (a) The factor VIII
- Page 538:
252 Figure 14.7 Part III The Applic
- Page 542:
254 Figure 14.8 Part III The Applic
- Page 546:
256 Part III The Applications of Ge
- Page 550:
258 Figure 14.10 Part III The Appli
- Page 554:
260 Figure 14.11 Differentiation of
- Page 558:
262 Part III The Applications of Ge
- Page 562:
264 Chapter 15 Gene Cloning and DNA
- Page 566:
266 Table 15.1 Part III The Applica
- Page 570:
268 Figure 15.3 Positional effects.
- Page 574:
270 (a) Production of two toxins (c
- Page 578:
272 (a) Detoxification of glyphosat
- Page 582:
274 are being produced by transferr
- Page 586:
276 Figure 15.10 The differences in
- Page 590:
278 Figure 15.11 DNA excision by th
- Page 594:
280 Part III The Applications of Ge
- Page 598:
282 Chapter 16 Gene Cloning and DNA
- Page 602:
284 Figure 16.1 Genetic fingerprint
- Page 606:
286 Figure 16.3 Inheritance of STR
- Page 610:
288 Figure 16.5 Part III The Applic
- Page 614:
290 Figure 16.6 Sex identification
- Page 618:
292 Figure 16.8 Part III The Applic
- Page 622:
294 Part III The Applications of Ge
- Page 626:
296 Figure 16.11 Origin of agricult
- Page 630:
298 Glossary GLOSSARY 2 Fm circle A
- Page 634:
300 Glossary Chromosome walking A t
- Page 638:
302 Glossary Ethanol precipitation
- Page 642:
304 Glossary In vitro mutagenesis A
- Page 646:
306 Glossary Nick translation The r
- Page 650:
308 Glossary Proteome The entire pr
- Page 654:
310 Glossary Single nucleotide poly
- Page 658:
312 INDEX Index (g = glossary, f =
- Page 662:
314 cloning vectors (continued) exp
- Page 666:
316 herpes simplex virus glycoprote
- Page 670:
318 polyethylene glycol, see PEG po
- Page 674:
320 T m , 153, 305g tobacco, 269 TO