- Page 4:
GENE CLONING AND DNA ANALYSIS
- Page 10:
This edition first published 2010,
- Page 16:
Contents CONTENTS Preface to the Si
- Page 20:
Contents ix 4.3 Ligation—joining
- Page 24:
Contents xi 8 How to Obtain a Clone
- Page 28:
Contents xiii 11.3 Identifying and
- Page 32:
Contents xv 15.1.2 Herbicide resist
- Page 36:
PART I The Basic Principles of Gene
- Page 42:
4 Part I The Basic Principles of Ge
- Page 46:
6 1.4 What is PCR? Figure 1.2 The b
- Page 50:
8 Figure 1.3 Cloning allows individ
- Page 54:
10 Figure 1.5 Gene isolation by PCR
- Page 58:
12 Further reading FURTHER READING
- Page 62:
14 Figure 2.1 Plasmids: independent
- Page 66:
16 Figure 2.4 Plasmid transfer by c
- Page 70:
18 Figure 2.6 Capsid components Pha
- Page 74:
20 Figure 2.7 λ phage particle att
- Page 78:
22 (a) The linear form of the λ DN
- Page 82:
24 Part I The Basic Principles of G
- Page 86:
26 Bacterial culture Table 3.1 Cent
- Page 90:
28 Bacterial culture Figure 3.3 Har
- Page 94:
30 Figure 3.6 Removal of protein co
- Page 98:
32 Figure 3.8 Collecting DNA by eth
- Page 102:
34 Figure 3.10 DNA purification by
- Page 106:
36 Figure 3.12 Two conformations of
- Page 110:
38 Figure 3.15 Partial unwinding of
- Page 114:
40 Figure 3.18 Preparation of a pha
- Page 118:
42 Figure 3.21 Collection of phage
- Page 122:
44 Further reading FURTHER READING
- Page 126:
46 Part I The Basic Principles of G
- Page 130:
48 Figure 4.3 The reactions catalyz
- Page 134:
50 Figure 4.6 (a) Alkaline phosphat
- Page 138:
52 Figure 4.8 (a) Restriction of ph
- Page 142:
54 Figure 4.9 (a) Production of blu
- Page 146:
56 Table 4.2 A 10 × buffer suitabl
- Page 150:
58 Figure 4.13 Visualizing DNA band
- Page 154:
60 Figure 4.15 Using a restriction
- Page 158:
62 Figure 4.17 The influence of DNA
- Page 162:
64 Figure 4.20 (a) Ligating blunt e
- Page 166:
66 Figure 4.23 Adaptors and the pot
- Page 170:
68 Figure 4.25 The use of adaptors:
- Page 174:
70 Figure 4.28 (a) The ends of the
- Page 178:
72 Chapter 5 Introduction of DNA in
- Page 182:
74 Part I The Basic Principles of G
- Page 186:
76 Figure 5.4 Normal E. coli cell (
- Page 190:
78 Figure 5.8 (b) Replica plating W
- Page 194:
80 Figure 5.10 (a) The role of the
- Page 198:
82 Figure 5.11 (a) A single-strain
- Page 202:
84 Figure 5.13 Strategies for the s
- Page 206:
86 Figure 5.14 Figure 5.15 (a) Prec
- Page 210:
88 Chapter 6 Cloning Vectors for E.
- Page 214:
90 during purification. pBR322 is 4
- Page 218:
92 (a) pUC8 (b) Restriction sites i
- Page 222:
94 Figure 6.5 The M13 genome, showi
- Page 226:
96 not totally disrupt the lacZ′
- Page 230:
98 Figure 6.10 The λ genetic map,
- Page 234:
100 (a) Cloning with a λ replaceme
- Page 238:
102 Figure 6.15 (a) A typical cosmi
- Page 242:
104 capsid structure. Cosmid-type v
- Page 246:
106 Figure 7.1 The yeast 2 µm plas
- Page 250:
108 Figure 7.4 Cloning with an E. c
- Page 254:
110 Figure 7.7 Chromosome structure
- Page 258:
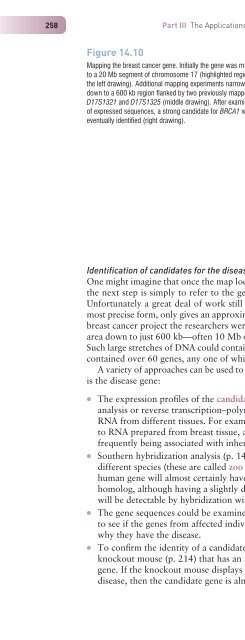
112 for instance to examine the seg
- Page 262:
114 Figure 7.10 The Ti plasmid and
- Page 266:
116 (a) Wound infection by recombin
- Page 270:
118 Figure 7.15 Direct gene transfe
- Page 274:
120 Caulimovirus vectors Although o
- Page 278:
122 Figure 7.18 Cloning in Drosophi
- Page 282:
124 cause the death of the mouse ce
- Page 286:
126 Chapter 8 How to Obtain a Clone
- Page 290:
128 Figure 8.2 The basic strategies
- Page 294:
130 Figure 8.4 (a) Construction of
- Page 298:
132 Figure 8.5 Preparation of a gen
- Page 302:
134 Figure 8.7 One possible scheme
- Page 306:
136 Figure 8.10 The structure of α
- Page 310:
138 Figure 8.12 Two methods for the
- Page 314:
140 Figure 8.15 A simplified scheme
- Page 318:
142 Figure 8.17 Heterologous probin
- Page 322:
144 skills (Figure 8.19b). The same
- Page 326:
146 The problem of gene expression
- Page 330:
148 Figure 9.1 Hybridization of the
- Page 334:
150 Figure 9.3 Figure 9.4 5’ 3’
- Page 338:
152 Figure 9.7 A typical temperatur
- Page 342:
154 Figure 9.9 Calculating the T m
- Page 346:
156 Figure 9.13 5’ 3’ A G A C T
- Page 350:
158 Figure 9.16 Double-stranded PCR
- Page 354:
160 Figure 9.18 Hybridization of a
- Page 360:
PART II The Applications of Gene Cl
- Page 366:
166 termination sequencing has gain
- Page 370:
168 Figure 10.3 Reading the sequenc
- Page 374:
170 Figure 10.5 The basis to therma
- Page 378:
172 Figure 10.7 Pyrosequencing. Fig
- Page 382:
174 Figure 10.9 Part II The Applica
- Page 386:
176 Figure 10.11 Using oligonucleot
- Page 390:
178 Figure 10.13 Chromosome walking
- Page 394:
180 Figure 10.16 Part II The Applic
- Page 398:
182 Figure 10.20 Fluorescent in sit
- Page 402:
184 s Part II The Applications of G
- Page 406:
186 Figure 11.1 Some fundamentals o
- Page 410:
188 Figure 11.3 The 5′ end of an
- Page 414:
190 Figure 11.6 5' 3' RNA transcrip
- Page 418:
192 Figure 11.8 Possible positions
- Page 422:
194 Figure 11.11 A bound protein pr
- Page 426:
196 Figure 11.14 A modification int
- Page 430:
198 Table 11.1 Figure 11.16 A repor
- Page 434:
200 Pure mRNA Labelled translation
- Page 438:
202 Figure 11.22 (a) Restriction fr
- Page 442:
204 Part II The Applications of Gen
- Page 446:
206 Part II The Applications of Gen
- Page 450:
208 as molecular biology in silico,
- Page 454:
210 Figure 12.4 Part II The Applica
- Page 458:
212 Figure 12.7 Using comparisons b
- Page 462:
214 Figure 12.10 The use of a yeast
- Page 466:
216 Figure 12.11 Serial analysis of
- Page 470:
218 Load the protein sample Figure
- Page 474:
220 Figure 12.16 Analyzing two prot
- Page 478:
222 Part II The Applications of Gen
- Page 484:
Chapter 13 Production of Protein fr
- Page 488:
Chapter 13 Production of Protein fr
- Page 492:
Chapter 13 Production of Protein fr
- Page 496:
Chapter 13 Production of Protein fr
- Page 500: Chapter 13 Production of Protein fr
- Page 504: Chapter 13 Production of Protein fr
- Page 508: Chapter 13 Production of Protein fr
- Page 512: Chapter 13 Production of Protein fr
- Page 516: Chapter 13 Production of Protein fr
- Page 520: Chapter 13 Production of Protein fr
- Page 524: Chapter 14 Gene Cloning and DNA Ana
- Page 528: Chapter 14 Gene Cloning and DNA Ana
- Page 532: Chapter 14 Gene Cloning and DNA Ana
- Page 536: Chapter 14 Gene Cloning and DNA Ana
- Page 540: Chapter 14 Gene Cloning and DNA Ana
- Page 544: Chapter 14 Gene Cloning and DNA Ana
- Page 548: Chapter 14 Gene Cloning and DNA Ana
- Page 554: 260 Figure 14.11 Differentiation of
- Page 558: 262 Part III The Applications of Ge
- Page 562: 264 Chapter 15 Gene Cloning and DNA
- Page 566: 266 Table 15.1 Part III The Applica
- Page 570: 268 Figure 15.3 Positional effects.
- Page 574: 270 (a) Production of two toxins (c
- Page 578: 272 (a) Detoxification of glyphosat
- Page 582: 274 are being produced by transferr
- Page 586: 276 Figure 15.10 The differences in
- Page 590: 278 Figure 15.11 DNA excision by th
- Page 594: 280 Part III The Applications of Ge
- Page 598: 282 Chapter 16 Gene Cloning and DNA
- Page 602:
284 Figure 16.1 Genetic fingerprint
- Page 606:
286 Figure 16.3 Inheritance of STR
- Page 610:
288 Figure 16.5 Part III The Applic
- Page 614:
290 Figure 16.6 Sex identification
- Page 618:
292 Figure 16.8 Part III The Applic
- Page 622:
294 Part III The Applications of Ge
- Page 626:
296 Figure 16.11 Origin of agricult
- Page 630:
298 Glossary GLOSSARY 2 Fm circle A
- Page 634:
300 Glossary Chromosome walking A t
- Page 638:
302 Glossary Ethanol precipitation
- Page 642:
304 Glossary In vitro mutagenesis A
- Page 646:
306 Glossary Nick translation The r
- Page 650:
308 Glossary Proteome The entire pr
- Page 654:
310 Glossary Single nucleotide poly
- Page 658:
312 INDEX Index (g = glossary, f =
- Page 662:
314 cloning vectors (continued) exp
- Page 666:
316 herpes simplex virus glycoprote
- Page 670:
318 polyethylene glycol, see PEG po
- Page 674:
320 T m , 153, 305g tobacco, 269 TO