Gene Cloning and DNA Analysis: An Introduction, Sixth Edition ...
Gene Cloning and DNA Analysis: An Introduction, Sixth Edition ...
Gene Cloning and DNA Analysis: An Introduction, Sixth Edition ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
166<br />
termination sequencing has gained pre-eminence for several reasons, not least being the<br />
relative ease with which the technique can be automated. As we will see later in this<br />
chapter, in order to sequence an entire genome a huge number of individual sequencing<br />
experiments must be carried out, <strong>and</strong> it would take many years to perform all of<br />
these by h<strong>and</strong>. Automated sequencing techniques are therefore essential if a genome<br />
project is to be completed in a reasonable timespan.<br />
Part of the automation strategy is to design systems that enable many individual<br />
sequencing experiments to be carried out at once. With the chain termination method,<br />
up to 96 sequences can be obtained simultaneously in a single run of a sequencing<br />
machine. This is still not enough to fully satisfy the dem<strong>and</strong>s of genome sequencing,<br />
<strong>and</strong> during the last few years an alternative method called pyrosequencing has become<br />
popular. Pyrosequencing, which was invented in 1998, forms the basis to a massively<br />
parallel strategy that enables hundreds of thous<strong>and</strong>s of short sequences to be generated<br />
at the same time.<br />
10.1.1 Chain termination <strong>DNA</strong> sequencing<br />
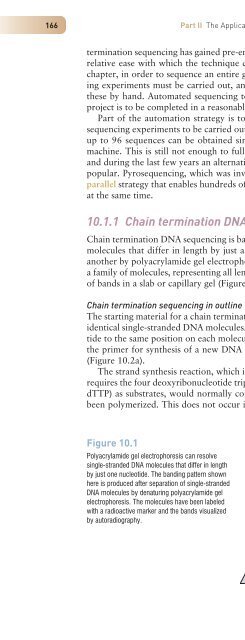
Chain termination <strong>DNA</strong> sequencing is based on the principle that single-str<strong>and</strong>ed <strong>DNA</strong><br />
molecules that differ in length by just a single nucleotide can be separated from one<br />
another by polyacrylamide gel electrophoresis. This means that it is possible to resolve<br />
a family of molecules, representing all lengths from 10 to 1500 nucleotides, into a series<br />
of b<strong>and</strong>s in a slab or capillary gel (Figure 10.1).<br />
Chain termination sequencing in outline<br />
The starting material for a chain termination sequencing experiment is a preparation of<br />
identical single-str<strong>and</strong>ed <strong>DNA</strong> molecules. The first step is to anneal a short oligonucleotide<br />
to the same position on each molecule, this oligonucleotide subsequently acting as<br />
the primer for synthesis of a new <strong>DNA</strong> str<strong>and</strong> that is complementary to the template<br />
(Figure 10.2a).<br />
The str<strong>and</strong> synthesis reaction, which is catalyzed by a <strong>DNA</strong> polymerase enzyme <strong>and</strong><br />
requires the four deoxyribonucleotide triphosphates (dNTPs—dATP, dCTP, dGTP, <strong>and</strong><br />
dTTP) as substrates, would normally continue until several thous<strong>and</strong> nucleotides had<br />
been polymerized. This does not occur in a chain termination sequencing experiment<br />
Figure 10.1<br />
Polyacrylamide gel electrophoresis can resolve<br />
single-str<strong>and</strong>ed <strong>DNA</strong> molecules that differ in length<br />
by just one nucleotide. The b<strong>and</strong>ing pattern shown<br />
here is produced after separation of single-str<strong>and</strong>ed<br />
<strong>DNA</strong> molecules by denaturing polyacrylamide gel<br />
electrophoresis. The molecules have been labeled<br />
with a radioactive marker <strong>and</strong> the b<strong>and</strong>s visualized<br />
by autoradiography.<br />
Part II The Applications of <strong>Gene</strong> <strong>Cloning</strong> <strong>and</strong> <strong>DNA</strong> <strong><strong>An</strong>alysis</strong> in Research<br />
10 nucleotides<br />
50 nucleotides