Comparative day/night metatranscriptomic analysis of microbial ...
Comparative day/night metatranscriptomic analysis of microbial ...
Comparative day/night metatranscriptomic analysis of microbial ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
% PHX Genes Frequency<br />
Frequency<br />
Based on these coverage estimates, increased<br />
sequencing depth would have been required to fully<br />
capture some specialized processes carried out by rarer<br />
members <strong>of</strong> the HOT community, but frequently transcribed<br />
genes from abundant taxa were well represented.<br />
In support <strong>of</strong> this, transcript mapping to the three P. mari-<br />
Number <strong>of</strong> Adjacent Genes<br />
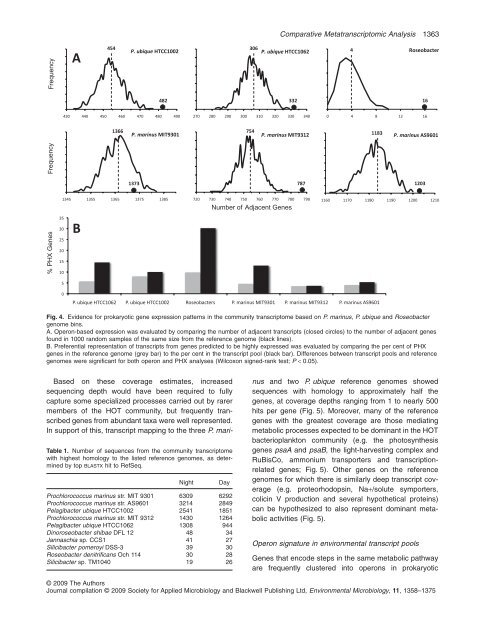
Fig. 4. Evidence for prokaryotic gene expression patterns in the community transcriptome based on P. marinus, P. ubique and Roseobacter<br />
genome bins.<br />
A. Operon-based expression was evaluated by comparing the number <strong>of</strong> adjacent transcripts (closed circles) to the number <strong>of</strong> adjacent genes<br />
found in 1000 random samples <strong>of</strong> the same size from the reference genome (black lines).<br />
B. Preferential representation <strong>of</strong> transcripts from genes predicted to be highly expressed was evaluated by comparing the per cent <strong>of</strong> PHX<br />
genes in the reference genome (grey bar) to the per cent in the transcript pool (black bar). Differences between transcript pools and reference<br />
genomes were significant for both operon and PHX analyses (Wilcoxon signed-rank test; P < 0.05).<br />
Table 1. Number <strong>of</strong> sequences from the community transcriptome<br />
with highest homology to the listed reference genomes, as determined<br />
by top BLASTX hit to RefSeq.<br />
Night Day<br />
Prochlorococcus marinus str. MIT 9301 6309 6292<br />
Prochlorococcus marinus str. AS9601 3214 2849<br />
Pelagibacter ubique HTCC1002 2541 1851<br />
Prochlorococcus marinus str. MIT 9312 1430 1264<br />
Pelagibacter ubique HTCC1062 1308 944<br />
Dinoroseobacter shibae DFL 12 48 34<br />
Jannaschia sp. CCS1 41 27<br />
Silicibacter pomeroyi DSS-3 39 30<br />
Roseobacter denitrificans Och 114 30 28<br />
Silicibacter sp. TM1040 19 26<br />
<strong>Comparative</strong> Metatranscriptomic Analysis 1363<br />
nus and two P. ubique reference genomes showed<br />
sequences with homology to approximately half the<br />
genes, at coverage depths ranging from 1 to nearly 500<br />
hits per gene (Fig. 5). Moreover, many <strong>of</strong> the reference<br />
genes with the greatest coverage are those mediating<br />
metabolic processes expected to be dominant in the HOT<br />
bacterioplankton community (e.g. the photosynthesis<br />
genes psaA and psaB, the light-harvesting complex and<br />
RuBisCo, ammonium transporters and transcriptionrelated<br />
genes; Fig. 5). Other genes on the reference<br />
genomes for which there is similarly deep transcript coverage<br />
(e.g. proteorhodopsin, Na+/solute symporters,<br />
colicin V production and several hypothetical proteins)<br />
can be hypothesized to also represent dominant metabolic<br />
activities (Fig. 5).<br />
Operon signature in environmental transcript pools<br />
Genes that encode steps in the same metabolic pathway<br />
are frequently clustered into operons in prokaryotic<br />
© 2009 The Authors<br />
Journal compilation © 2009 Society for Applied Microbiology and Blackwell Publishing Ltd, Environmental Microbiology, 11, 1358–1375