Comparative day/night metatranscriptomic analysis of microbial ...
Comparative day/night metatranscriptomic analysis of microbial ...
Comparative day/night metatranscriptomic analysis of microbial ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
1366 R. S. Poretsky et al.<br />
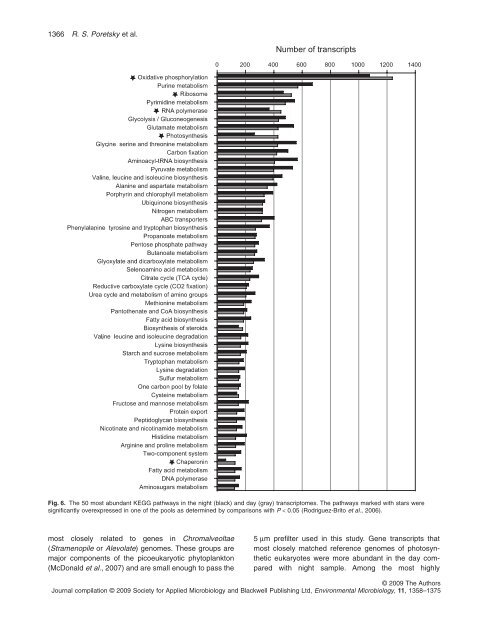
Fig. 6. The 50 most abundant KEGG pathways in the <strong>night</strong> (black) and <strong>day</strong> (gray) transcriptomes. The pathways marked with stars were<br />
significantly overexpressed in one <strong>of</strong> the pools as determined by comparisons with P < 0.05 (Rodriguez-Brito et al., 2006).<br />
most closely related to genes in Chromalveoltae<br />
(Stramenopile or Alevolate) genomes. These groups are<br />
major components <strong>of</strong> the picoeukaryotic phytoplankton<br />
(McDonald et al., 2007) and are small enough to pass the<br />
5 mm prefilter used in this study. Gene transcripts that<br />
most closely matched reference genomes <strong>of</strong> photosynthetic<br />
eukaryotes were more abundant in the <strong>day</strong> compared<br />
with <strong>night</strong> sample. Among the most highly<br />
© 2009 The Authors<br />
Journal compilation © 2009 Society for Applied Microbiology and Blackwell Publishing Ltd, Environmental Microbiology, 11, 1358–1375