Undergraduate Research Journal
Undergraduate Research Journal
Undergraduate Research Journal
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Molecular Analysis of the Interaction Between Staphylococcus aureus Protein Sbi and Immune System Protein C3d<br />
Wilson Rodriguez<br />
Introduction<br />
The complement system is part of our innate immune<br />
system and delivers potent responses to pathogenic<br />
infection and disease. Complement component 3 (C3)<br />
serves as the central protein of the complement system<br />
and can be activated by the classical, lectin, and alternative<br />
pathways. Cleavage of C3 results in fragments C3a and<br />
C3b, which can participate in amplification of immune<br />
response through the formation of convertases, as well as<br />
proinflamatory responses, chemotaxis, and the formation<br />
of the membrane attack complex (MAC). From these<br />
fragments, C3b tags pathogens, through the formation of a<br />
thioester bond, for subsequent recognition and elimination<br />
by macrophages. C3a interacts with C3aR on the surfaces of<br />
leukocytes and induces cell mobilization and chemotaxis.<br />
MAC is responsible for directly killing pathogens by cell<br />
lysis. 9 Simultaneous interactions with factors H and I lead<br />
to cleavage of C3b, yielding iC3b (inactive C3b) and C3d,<br />
with both fragments containing the thioeter bond moiety.<br />
C3d interacts with Complement Receptor 2 (CR2) and<br />
antigen-antibody complexes to facilitate B- and T- cell<br />
immune response. 7 Through the C3d fragment and the<br />
formation of the B cell receptor-coreceptor complex, the<br />
complement system acts as a link between innate and<br />
adaptive immunities by augmenting antibody responses.<br />
the C3d-CR2 complex from forming and thus blocking C3d<br />
from interacting with other branches of the host’s immune<br />
system such as the B and T cells. It is also hypothesized<br />
that Sbi interacts with residues involved in thioester<br />
bond formation between C3d and bacterial surfaces, thus<br />
inhibiting binding to pathogenic cell surfaces and retarding<br />
the formation of convertases in the classical pathway. 3<br />
In this study we analyze the effects of mutant variants,<br />
which are predictive of protein binding and stability.<br />
Through the generation of mutant variants and the<br />
calculation of electrostatic potentials, we can elucidate<br />
residues critical to the binding mechanism. 1 With these<br />
computational mutagenesis results we can model and<br />
design therapeutics tailored to enhance or inhibit specific<br />
protein-protein interactions.<br />
Although our immune responses are able to combat most<br />
infections, most bacterial pathogens have evolved and<br />
adapted methods for evading the human complement<br />
immune system. S. aureus in particular has developed a<br />
variety of immuno-regulatory proteins for the goal of<br />
immune evasion and does so by regulating the innate and<br />
adaptive immune systems. S. aureus also has the ability to<br />
control host regulatory proteins, thus facilitating bacterial<br />
function within the host. 7 One recently identified protein<br />
responsible for a variety of immune evasive functions<br />
is S. aureus binder of immunoglobulin G (Sbi). Sbi<br />
affects the adaptive immune system by binding to host<br />
immunoglobulin G3, but Sbi can also affect the alternative<br />
pathway of complement by interacting with C3d. Several<br />
experiments revealed that Sbi binds to C3d in a one-toone<br />
ratio but can bind to C3d at two locations: Complex 1 and<br />
Complex 23. This mode of binding allows Sbi to potently<br />
counter the complement activation. In Complex 1, Sbi inhibits<br />
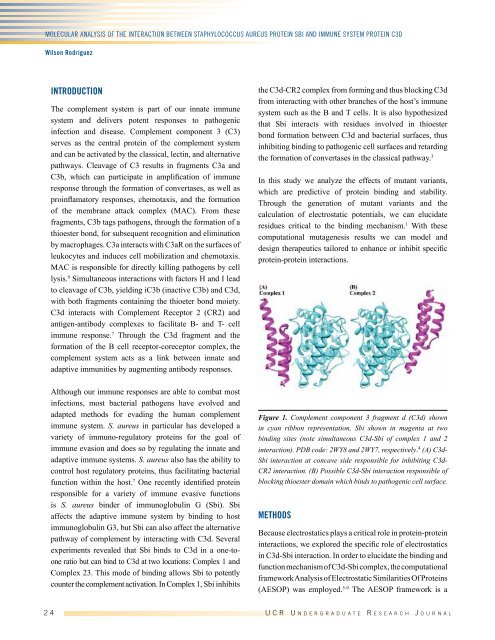
Figure 1. Complement component 3 fragment d (C3d) shown<br />
in cyan ribbon representation, Sbi shown in magenta at two<br />
binding sites (note simultaneous C3d-Sbi of complex 1 and 2<br />
interaction). PDB code: 2WY8 and 2WY7, respectively. 4 (A) C3d-<br />
Sbi interaction at concave side responsible for inhibiting C3d-<br />
CR2 interaction. (B) Possible C3d-Sbi interaction responsible of<br />
blocking thioester domain which binds to pathogenic cell surface.<br />
Methods<br />
Because electrostatics plays a critical role in protein-protein<br />
interactions, we explored the specific role of electrostatics<br />
in C3d-Sbi interaction. In order to elucidate the binding and<br />
function mechanism of C3d-Sbi complex, the computational<br />
framework Analysis of Electrostatic Similarities Of Proteins<br />
(AESOP) was employed. 5-9 The AESOP framework is a<br />
2 4 U C R U n d e r g r a d u a t e R e s e a r c h J o u r n a l