Undergraduate Research Journal
Undergraduate Research Journal
Undergraduate Research Journal
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Molecular Analysis of the Interaction Between Staphylococcus aureus Protein Sbi and Immune System Protein C3d<br />
Wilson Rodriguez<br />
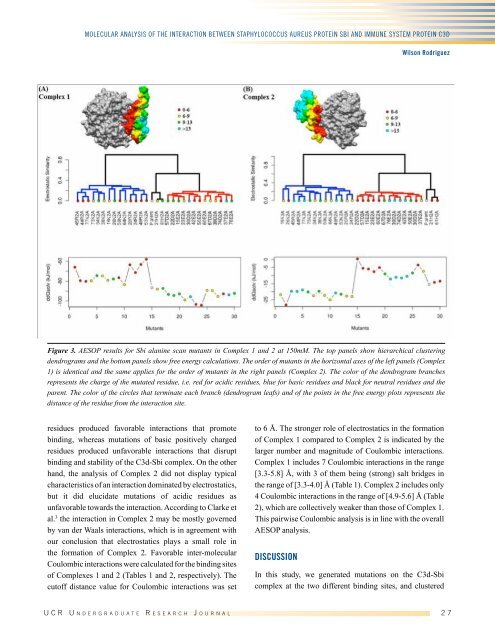
Figure 3. AESOP results for Sbi alanine scan mutants in Complex 1 and 2 at 150mM. The top panels show hierarchical clustering<br />
dendrograms and the bottom panels show free energy calculations. The order of mutants in the horizontal axes of the left panels (Complex<br />
1) is identical and the same applies for the order of mutants in the right panels (Complex 2). The color of the dendrogram branches<br />
represents the charge of the mutated residue, i.e. red for acidic residues, blue for basic residues and black for neutral residues and the<br />
parent. The color of the circles that terminate each branch (dendrogram leafs) and of the points in the free energy plots represents the<br />
distance of the residue from the interaction site.<br />
residues produced favorable interactions that promote<br />
binding, whereas mutations of basic positively charged<br />
residues produced unfavorable interactions that disrupt<br />
binding and stability of the C3d-Sbi complex. On the other<br />
hand, the analysis of Complex 2 did not display typical<br />
characteristics of an interaction dominated by electrostatics,<br />
but it did elucidate mutations of acidic residues as<br />
unfavorable towards the interaction. According to Clarke et<br />
al. 3 the interaction in Complex 2 may be mostly governed<br />
by van der Waals interactions, which is in agreement with<br />
our conclusion that electrostatics plays a small role in<br />
the formation of Complex 2. Favorable inter-molecular<br />
Coulombic interactions were calculated for the binding sites<br />
of Complexes 1 and 2 (Tables 1 and 2, respectively). The<br />
cutoff distance value for Coulombic interactions was set<br />
to 6 Å. The stronger role of electrostatics in the formation<br />
of Complex 1 compared to Complex 2 is indicated by the<br />
larger number and magnitude of Coulombic interactions.<br />
Complex 1 includes 7 Coulombic interactions in the range<br />
[3.3-5.8] Å, with 3 of them being (strong) salt bridges in<br />
the range of [3.3-4.0] Å (Table 1). Complex 2 includes only<br />
4 Coulombic interactions in the range of [4.9-5.6] Å (Table<br />
2), which are collectively weaker than those of Complex 1.<br />
This pairwise Coulombic analysis is in line with the overall<br />
AESOP analysis.<br />
DISCUSSION<br />
In this study, we generated mutations on the C3d-Sbi<br />
complex at the two different binding sites, and clustered<br />
U C R U n d e r g r a d u a t e R e s e a r c h J o u r n a l 2 7