in vivo
in vivo
in vivo
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Makio Tokunaga<br />
Look<strong>in</strong>g beneath the surface<br />
A new imag<strong>in</strong>g technique allows scientists to effectively visualize <strong>in</strong>dividual<br />
molecules with<strong>in</strong> liv<strong>in</strong>g cells <strong>in</strong> real-time<br />
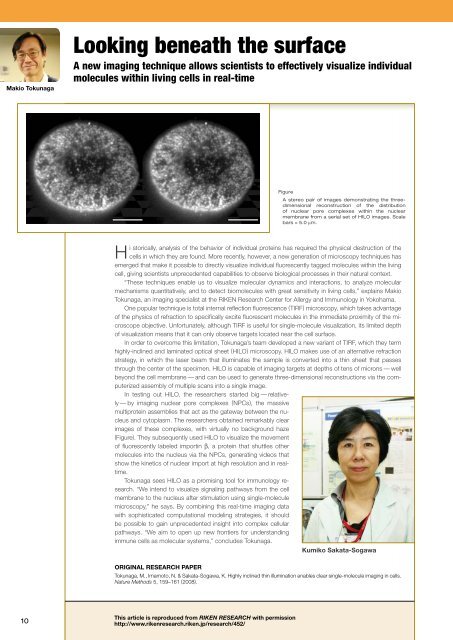
Figure<br />
A stereo pair of images demonstrat<strong>in</strong>g the threedimensional<br />
reconstruction of the distribution<br />
of nuclear pore complexes with<strong>in</strong> the nuclear<br />
membrane from a serial set of HILO images. Scale<br />
bars = 5.0 μm.<br />
Hi storically, analysis of the behavior of <strong>in</strong>dividual prote<strong>in</strong>s has required the physical destruction of the<br />
cells <strong>in</strong> which they are found. More recently, however, a new generation of microscopy techniques has<br />
emerged that make it possible to directly visualize <strong>in</strong>dividual fluorescently tagged molecules with<strong>in</strong> the liv<strong>in</strong>g<br />
cell, giv<strong>in</strong>g scientists unprecedented capabilities to observe biological processes <strong>in</strong> their natural context.<br />
“These techniques enable us to visualize molecular dynamics and <strong>in</strong>teractions, to analyze molecular<br />
mechanisms quantitatively, and to detect biomolecules with great sensitivity <strong>in</strong> liv<strong>in</strong>g cells,” expla<strong>in</strong>s Makio<br />
Tokunaga, an imag<strong>in</strong>g specialist at the RIKEN Research Center for Allergy and Immunology <strong>in</strong> Yokohama.<br />
One popular technique is total <strong>in</strong>ternal reflection fluorescence (TIRF) microscopy, which takes advantage<br />
of the physics of refraction to specifically excite fluorescent molecules <strong>in</strong> the immediate proximity of the microscope<br />
objective. Unfortunately, although TIRF is useful for s<strong>in</strong>gle-molecule visualization, its limited depth<br />
of visualization means that it can only observe targets located near the cell surface.<br />
In order to overcome this limitation, Tokunaga’s team developed a new variant of TIRF, which they term<br />
highly-<strong>in</strong>cl<strong>in</strong>ed and lam<strong>in</strong>ated optical sheet (HILO) microscopy. HILO makes use of an alternative refraction<br />
strategy, <strong>in</strong> which the laser beam that illum<strong>in</strong>ates the sample is converted <strong>in</strong>to a th<strong>in</strong> sheet that passes<br />
through the center of the specimen. HILO is capable of imag<strong>in</strong>g targets at depths of tens of microns — well<br />
beyond the cell membrane — and can be used to generate three-dimensional reconstructions via the computerized<br />
assembly of multiple scans <strong>in</strong>to a s<strong>in</strong>gle image.<br />
In test<strong>in</strong>g out HILO, the researchers started big — relatively<br />
— by imag<strong>in</strong>g nuclear pore complexes (NPCs), the massive<br />
multiprote<strong>in</strong> assemblies that act as the gateway between the nucleus<br />
and cytoplasm. The researchers obta<strong>in</strong>ed remarkably clear<br />
images of these complexes, with virtually no background haze<br />
(Figure). They subsequently used HILO to visualize the movement<br />
of fluorescently labeled import<strong>in</strong> β, a prote<strong>in</strong> that shuttles other<br />
molecules <strong>in</strong>to the nucleus via the NPCs, generat<strong>in</strong>g videos that<br />
show the k<strong>in</strong>etics of nuclear import at high resolution and <strong>in</strong> realtime.<br />
Tokunaga sees HILO as a promis<strong>in</strong>g tool for immunology research.<br />
“We <strong>in</strong>tend to visualize signal<strong>in</strong>g pathways from the cell<br />
membrane to the nucleus after stimulation us<strong>in</strong>g s<strong>in</strong>gle-molecule<br />
microscopy,” he says. By comb<strong>in</strong><strong>in</strong>g this real-time imag<strong>in</strong>g data<br />
with sophisticated computational model<strong>in</strong>g strategies, it should<br />
be possible to ga<strong>in</strong> unprecedented <strong>in</strong>sight <strong>in</strong>to complex cellular<br />
pathways. “We aim to open up new frontiers for understand<strong>in</strong>g<br />
immune cells as molecular systems,” concludes Tokunaga.<br />
Kumiko Sakata-Sogawa<br />
ORIGINAL RESEARCH PAPER<br />
Tokunaga, M., Imamoto, N. & Sakata-Sogawa, K. Highly <strong>in</strong>cl<strong>in</strong>ed th<strong>in</strong> illum<strong>in</strong>ation enables clear s<strong>in</strong>gle-molecule imag<strong>in</strong>g <strong>in</strong> cells.<br />
Nature Methods 5, 159–161 (2008).<br />
10<br />
This article is reproduced from RIKEN RESEARCH with permission<br />
http://www.rikenresearch.riken.jp/research/452/