FIAS Scientific Report 2011 - Frankfurt Institute for Advanced Studies ...
FIAS Scientific Report 2011 - Frankfurt Institute for Advanced Studies ...
FIAS Scientific Report 2011 - Frankfurt Institute for Advanced Studies ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
DNA unzipping<br />
Collaborators: S.N. Volkov 1 , E.V. Paramonova 2 , A.V. Yakubovich 3 , A.V. Solov’yov 3<br />
1 Bogolyubov <strong>Institute</strong> <strong>for</strong> Theoretical Physics, Ukraine, 2 <strong>Institute</strong> of Mathematical Problems of Biology RAS, Russia,<br />
3 <strong>Frankfurt</strong> <strong>Institute</strong> <strong>for</strong> <strong>Advanced</strong> <strong>Studies</strong><br />
Short description:<br />
All-atom molecular dynamics (MD) simulations of DNA duplex unzipping in a water environment were per<strong>for</strong>med.<br />
The investigated DNA double helix consists of a Drew-Dickerson dodecamer sequence and a hairpin<br />
(AAG) attached to the end of the double-helix chain. The considered system is used to examine the process<br />
of DNA strand separation under the action of an external <strong>for</strong>ce. This process occurs in vivo and now is being<br />
intensively investigated in experiments with single molecules.<br />
Main results:<br />
• The DNA dodecamer duplex is consequently unzipped<br />
pair by pair by means of the steered MD. The unzipping<br />
trajectories turn out to be similar <strong>for</strong> the duplex parts with<br />
G·C content and rather distinct <strong>for</strong> the parts with A·T content.<br />
• It is shown that during the unzipping each pair experiences<br />
two types of motion: relatively quick rotation together<br />
with all the duplex and slower motion in the frame<br />
of the unzipping <strong>for</strong>k. In the course of opening, the complementary<br />
pair passes through several distinct states: (i)<br />
the closed state in the double helix, (ii) the metastable preopened<br />
state in the unzipping <strong>for</strong>k and (iii) the unbound<br />
state.<br />
• The per<strong>for</strong>med simulations show that water molecules<br />
participate in the stabilization of the metastable states of<br />
the preopened base pairs in the DNA unzipping <strong>for</strong>k.<br />
Related publications in <strong>2011</strong>:<br />
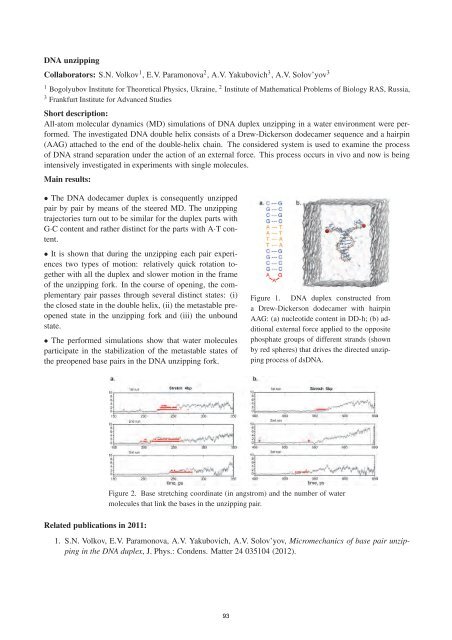
Figure 1. DNA duplex constructed from<br />
a Drew-Dickerson dodecamer with hairpin<br />
AAG: (a) nucleotide content in DD-h; (b) additional<br />
external <strong>for</strong>ce applied to the opposite<br />
phosphate groups of different strands (shown<br />
by red spheres) that drives the directed unzipping<br />
process of dsDNA.<br />
Figure 2. Base stretching coordinate (in angstrom) and the number of water<br />
molecules that link the bases in the unzipping pair.<br />
1. S.N. Volkov, E.V. Paramonova, A.V. Yakubovich, A.V. Solov’yov, Micromechanics of base pair unzipping<br />
in the DNA duplex, J. Phys.: Condens. Matter 24 035104 (2012).<br />
93