Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
2232 R. P. Verma, C. Hansch / Bioorg. Med. Chem. 15 (2007) 2223–2268<br />
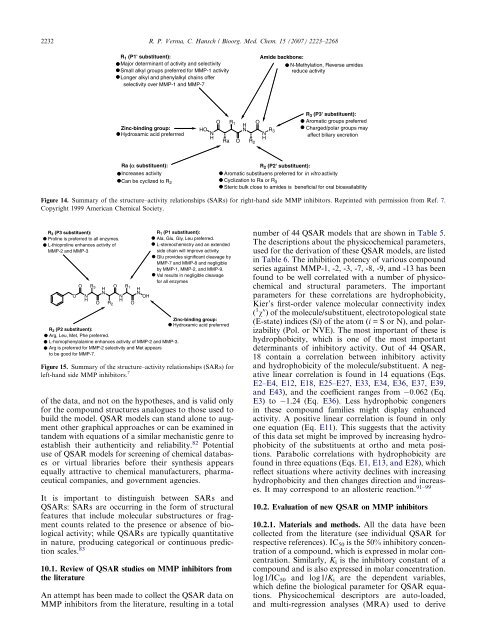
R 1 (P1' substituent):<br />
Major determinant of activity and selectivity<br />
Small alkyl groups preferred for MMP-1 activity<br />
Longer alkyl and phenylalkyl chains offer<br />
selectivity over MMP-1 and MMP-7<br />
Zinc-binding group:<br />
Hydroxamic acid preferrred<br />
Ra (α substituent):<br />
Increases activity<br />
Can be cyclized to R 2<br />
HO NH<br />
of the data, and not on the hypotheses, and is valid only<br />
for the compound structures analogues to those used to<br />
build the model. QSAR models can stand alone to augment<br />
other graphical approaches or can be examined in<br />
tandem with equations of a similar mechanistic genre to<br />
establish their authenticity and reliability. 82 Potential<br />
use of QSAR models for screening of chemical databases<br />
or virtual libraries before their synthesis appears<br />
equally attractive to chemical manufacturers, pharmaceutical<br />
companies, and government agencies.<br />
It is important to distinguish between SARs and<br />
QSARs: SARs are occurring in the form of structural<br />
features that include molecular substructures or fragment<br />
counts related to the presence or absence of biological<br />
activity; while QSARs are typically quantitative<br />
in nature, producing categorical or continuous prediction<br />
scales. 83<br />
10.1. Review of QSAR studies on MMP inhibitors from<br />
the literature<br />
An attempt has been made to collect the QSAR data on<br />
MMP inhibitors from the literature, resulting in a total<br />
O<br />
R 1<br />
H<br />
N<br />
Ra O R2 Amide backbone:<br />
O<br />
R3 N<br />
H<br />
N-Methylation, Reverse amides<br />
reduce activity<br />
R 3 (P3' substituent):<br />
Aromatic groups preferred<br />
Charged/polar groups may<br />
affect biliary excretion<br />
R2 (P2' substituent):<br />
Aromatic substituens preferred for in vitro activity<br />
Cyclization to Ra or R3 Steric bulk close to amides is beneficial for oral bioavailability<br />
Figure 14. Summary of the structure–activity relationships (SARs) for right-hand side MMP inhibitors. Reprinted with permission from Ref. 7.<br />
Copyright 1999 American Chemical Society.<br />
R 3 (P3 substituent):<br />
Proline is preferred to all enzymes.<br />
L-thioproline enhances activity of<br />
MMP-2 and MMP-3<br />
O<br />
O N H<br />
R 3<br />
O<br />
H<br />
N<br />
R 2<br />
O<br />
N<br />
H<br />
R 1<br />
O<br />
H<br />
N OH<br />
R 1 (P1 substituent):<br />
Ala, Glu, Gly, Leu preferred.<br />
L-stereochemistry and an extended<br />
side chain will improve activity.<br />
Glu provides significant cleavage by<br />
MMP-7 and MMP-8 and negligible<br />
by MMP-1, MMP-2, and MMP-9.<br />
Val results in negligible cleavage<br />
for all enzymes<br />
Zinc-binding group:<br />
Hydroxamic acid preferrred<br />
R2 (P2 substituent):<br />
Arg, Leu, Met, Phe preferred.<br />
L-homophenylalanine enhances activity of MMP-2 and MMP-3.<br />
Arg is preferred for MMP-2 selectivity and Met appears<br />
to be good for MMP-7.<br />
Figure 15. Summary of the structure–activity relationships (SARs) for<br />
left-hand side MMP inhibitors. 7<br />
number of 44 QSAR models that are shown in Table 5.<br />
The descriptions about the physicochemical parameters,<br />
used for the derivation of these QSAR models, are listed<br />
in Table 6. The inhibition potency of various compound<br />
series against MMP-1, -2, -3, -7, -8, -9, and -13 has been<br />
found to be well correlated with a number of physicochemical<br />
and structural parameters. The important<br />
parameters for these correlations are hydrophobicity,<br />
Kier’s first-order valence molecular connectivity index<br />
( 1 v v ) of the molecule/substituent, electrotopological state<br />
(E-state) indices (Si) of the atom (i = S or N), and polarizability<br />
(Pol. or NVE). The most important of these is<br />
hydrophobicity, which is one of the most important<br />
determinants of inhibitory activity. Out of 44 QSAR,<br />
18 contain a correlation between inhibitory activity<br />
and hydrophobicity of the molecule/substituent. A negative<br />
linear correlation is found in 14 equations (Eqs.<br />
E2–E4, E12, E18, E25–E27, E33, E34, E36, E37, E39,<br />
and E43), and the coefficient ranges from 0.062 (Eq.<br />
E3) to 1.24 (Eq. E36). Less hydrophobic congeners<br />
in these compound families might display enhanced<br />
activity. A positive linear correlation is found in only<br />
one equation (Eq. E11). This suggests that the activity<br />
of this data set might be improved by increasing hydrophobicity<br />
of the substituents at ortho and meta positions.<br />
Parabolic correlations with hydrophobicity are<br />
found in three equations (Eqs. E1, E13, and E28), which<br />
reflect situations where activity declines with increasing<br />
hydrophobicity and then changes direction and increases.<br />
It may correspond to an allosteric reaction. 91–99<br />
10.2. Evaluation of new QSAR on MMP inhibitors<br />
10.2.1. Materials and methods. All the data have been<br />
collected from the literature (see individual QSAR for<br />
respective references). IC50 is the 50% inhibitory concentration<br />
of a compound, which is expressed in molar concentration.<br />
Similarly, K i is the inhibitory constant of a<br />
compound and is also expressed in molar concentration.<br />
log1/IC50 and log1/Ki are the dependent variables,<br />
which define the biological parameter for QSAR equations.<br />
Physicochemical descriptors are auto-loaded,<br />
and multi-regression analyses (MRA) used to derive