Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
2252 R. P. Verma, C. Hansch / Bioorg. Med. Chem. 15 (2007) 2223–2268<br />
Table 29. Biological and physicochemical parameters used to derive QSAR equation 25 for the inhibition of MMP-3 by thiazepine derivatives (XXII)<br />
No. X Y log1/IC50 (Eq. 25) r +<br />
Obsd. Pred. D<br />
1 a<br />
S 4-OCH3 9.15 8.27 0.88 0.78<br />
2<br />
3<br />
SO24-OCH3 8.16 8.27 0.11 0.78<br />
a<br />
S 4-Br 8.00 6.90 1.10 0.15<br />
4 SO24-Br 7.05 6.90 0.15 0.15<br />
5 S 2-CH3,4-Br 7.06 7.36 0.30 0.16<br />
6 S 4-OC4H9 8.18 8.32 0.14 0.81<br />
7 SO24-OC4H9 8.57 8.32 0.25 0.81<br />
8 S 4-C3H7 7.34 7.55 0.21 0.29<br />
9 S 4-C5H11 7.60 7.55 0.05 0.29<br />
10 S 4-O(CH2)2OCH3 7.77 7.55 0.22 0.29<br />
11 S 4-OC6H5 7.96 7.86 0.10 0.50<br />
a Not included in the derivation of QSAR 25.<br />
a hydrogen bond between Leu-164 and the sulfonamide<br />
oxygen may develop.<br />
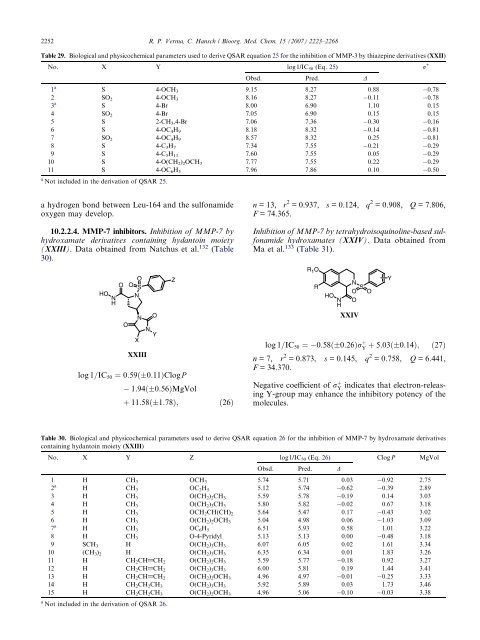
10.2.2.4. MMP-7 inhibitors. Inhibition of MMP-7 by<br />
hydroxamate derivatives containing hydantoin moiety<br />
(XXIII). Data obtained from Natchus et al. 132 (Table<br />
30).<br />
HO<br />
N<br />
H<br />
O<br />
O<br />
O<br />
O S<br />
N<br />
X<br />
N<br />
XXIII<br />
O<br />
N<br />
Y<br />
log 1=IC50 ¼ 0:59ð 0:11ÞClogP<br />
Z<br />
1:94ð 0:56ÞMgVol<br />
þ 11:58ð 1:78Þ; ð26Þ<br />
n = 13, r 2 = 0.937, s = 0.124, q 2 = 0.908, Q = 7.806,<br />
F = 74.365.<br />
Inhibition of MMP-7 by tetrahydroisoquinoline-based sulfonamide<br />
hydroxamates (XXIV). Data obtained from<br />
Ma et al. 133 (Table 31).<br />
R 1O<br />
R<br />
HO<br />
N<br />
H<br />
N<br />
S<br />
O O<br />
O<br />
XXIV<br />
log 1=IC50 ¼ 0:58ð 0:26Þr þ<br />
Y þ 5:03ð 0:14Þ; ð27Þ<br />
n =7, r 2 = 0.873, s = 0.145, q 2 = 0.758, Q = 6.441,<br />
F = 34.370.<br />
Negative coefficient of rþ Y indicates that electron-releasing<br />
Y-group may enhance the inhibitory potency of the<br />
molecules.<br />
Table 30. Biological and physicochemical parameters used to derive QSAR equation 26 for the inhibition of MMP-7 by hydroxamate derivatives<br />
containing hydantoin moiety (XXIII)<br />
No. X Y Z log1/IC50 (Eq. 26) ClogP MgVol<br />
Obsd. Pred. D<br />
1<br />
2<br />
H CH3OCH3 5.74 5.71 0.03 0.92 2.75<br />
a<br />
H CH3OC2H5 5.12 5.74 0.62 0.39 2.89<br />
3 H CH3O(CH2) 2CH3 5.59 5.78 0.19 0.14 3.03<br />
4 H CH3O(CH2)3CH3 5.80 5.82 0.02 0.67 3.18<br />
5 H CH3OCH2CH(CH)2 5.64 5.47 0.17 0.43 3.02<br />
6<br />
7<br />
H CH3O(CH2) 2OCH3 5.04 4.98 0.06 1.03 3.09<br />
a<br />
H CH3OC6H5 6.51 5.93 0.58 1.01 3.22<br />
8 H CH3O-4-Pyridyl 5.13 5.13 0.00 0.48 3.18<br />
9 SCH3 H O(CH2)3CH3 6.07 6.05 0.02 1.61 3.34<br />
10 (CH3) 2 H O(CH2) 3CH3 6.35 6.34 0.01 1.83 3.26<br />
11 H CH2CH@CH2 O(CH2)2CH3 5.59 5.77 0.18 0.92 3.27<br />
12 H CH2CH@CH2 O(CH2)3CH3 6.00 5.81 0.19 1.44 3.41<br />
13 H CH2CH@CH2 O(CH2)2OCH3 4.96 4.97 0.01 0.25 3.33<br />
14 H CH2CH2CH3 O(CH2) 3CH3 5.92 5.89 0.03 1.73 3.46<br />
15 H CH2CH2CH3 O(CH2)2OCH3 4.96 5.06 0.10 0.03 3.38<br />
a Not included in the derivation of QSAR 26.<br />
Y