Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
Matrix metalloproteinases (MMPs): Chemical–biological functions ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
2246 R. P. Verma, C. Hansch / Bioorg. Med. Chem. 15 (2007) 2223–2268<br />
Y<br />
X<br />
O<br />
H<br />
N<br />
XIII<br />
log 1=IC50 ¼ 0:35ð 0:16ÞClogP 3:63ð 0:68ÞI Y<br />
þ 7:61ð 0:60Þ; ð13Þ<br />
n =16, r 2 = 0.919, s = 0.471, q 2 = 0.869, Q = 2.036,<br />
F = 73.747.<br />
Positive ClogP suggests that highly hydrophobic P1 0<br />
modified phenylalanine analogues (XIII) would be more<br />
active. The indicator variable (I Y) applies to those compounds,<br />
which have Y = COOH. The negative coefficient<br />
of the indicator variable indicates that the<br />
presence of CONHOH group at Y-position will improve<br />
the activity. As the large coefficient of indicator variable<br />
(IY) suggests that compounds with Y = COOH and<br />
CONHOH may act by different mechanism.<br />
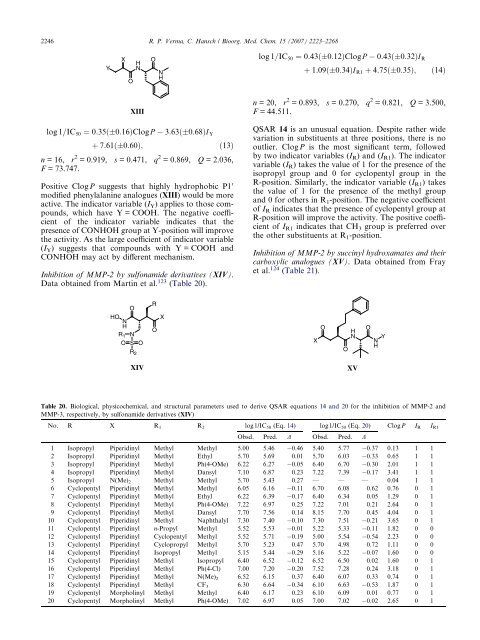
Inhibition of MMP-2 by sulfonamide derivatives (XIV).<br />
Data obtained from Martin et al. 123 (Table 20).<br />
HO NH<br />
O<br />
R1 N<br />
O S O<br />
R2 XIV<br />
O<br />
R<br />
O<br />
N<br />
H<br />
X<br />
log 1=IC50 ¼ 0:43ð 0:12ÞClogP 0:43ð 0:32ÞI R<br />
þ 1:09ð 0:34ÞI R1 þ 4:75ð 0:35Þ; ð14Þ<br />
n = 20, r 2 = 0.893, s = 0.270, q 2 = 0.821, Q = 3.500,<br />
F = 44.511.<br />
QSAR 14 is an unusual equation. Despite rather wide<br />
variation in substituents at three positions, there is no<br />
outlier. ClogP is the most significant term, followed<br />
by two indicator variables (IR) and (IR1). The indicator<br />
variable (IR) takes the value of 1 for the presence of the<br />
isopropyl group and 0 for cyclopentyl group in the<br />
R-position. Similarly, the indicator variable (IR1) takes<br />
the value of 1 for the presence of the methyl group<br />
and 0 for others in R 1-position. The negative coefficient<br />
of IR indicates that the presence of cyclopentyl group at<br />
R-position will improve the activity. The positive coefficient<br />
of I R1 indicates that CH 3 group is preferred over<br />
the other substituents at R1-position.<br />
Inhibition of MMP-2 by succinyl hydroxamates and their<br />
carboxylic analogues (XV). Data obtained from Fray<br />
et al. 124 (Table 21).<br />
Table 20. Biological, physicochemical, and structural parameters used to derive QSAR equations 14 and 20 for the inhibition of MMP-2 and<br />
MMP-3, respectively, by sulfonamide derivatives (XIV)<br />
No. R X R1 R2 log1/IC50 (Eq. 14) log1/IC50 (Eq. 20) ClogP IR IR1<br />
Obsd. Pred. D Obsd. Pred. D<br />
1 Isopropyl Piperidinyl Methyl Methyl 5.00 5.46 0.46 5.40 5.77 0.37 0.13 1 1<br />
2 Isopropyl Piperidinyl Methyl Ethyl 5.70 5.69 0.01 5.70 6.03 0.33 0.65 1 1<br />
3 Isopropyl Piperidinyl Methyl Ph(4-OMe) 6.22 6.27 0.05 6.40 6.70 0.30 2.01 1 1<br />
4 Isopropyl Piperidinyl Methyl Dansyl 7.10 6.87 0.23 7.22 7.39 0.17 3.41 1 1<br />
5 Isopropyl N(Me)2 Methyl Methyl 5.70 5.43 0.27 — — — 0.04 1 1<br />
6 Cyclopentyl Piperidinyl Methyl Methyl 6.05 6.16 0.11 6.70 6.08 0.62 0.76 0 1<br />
7 Cyclopentyl Piperidinyl Methyl Ethyl 6.22 6.39 0.17 6.40 6.34 0.05 1.29 0 1<br />
8 Cyclopentyl Piperidinyl Methyl Ph(4-OMe) 7.22 6.97 0.25 7.22 7.01 0.21 2.64 0 1<br />
9 Cyclopentyl Piperidinyl Methyl Dansyl 7.70 7.56 0.14 8.15 7.70 0.45 4.04 0 1<br />
10 Cyclopentyl Piperidinyl Methyl Naphthalyl 7.30 7.40 0.10 7.30 7.51 0.21 3.65 0 1<br />
11 Cyclopentyl Piperidinyl n-Propyl Methyl 5.52 5.53 0.01 5.22 5.33 0.11 1.82 0 0<br />
12 Cyclopentyl Piperidinyl Cyclopentyl Methyl 5.52 5.71 0.19 5.00 5.54 0.54 2.23 0 0<br />
13 Cyclopentyl Piperidinyl Cyclopropyl Methyl 5.70 5.23 0.47 5.70 4.98 0.72 1.11 0 0<br />
14 Cyclopentyl Piperidinyl Isopropyl Methyl 5.15 5.44 0.29 5.16 5.22 0.07 1.60 0 0<br />
15 Cyclopentyl Piperidinyl Methyl Isopropyl 6.40 6.52 0.12 6.52 6.50 0.02 1.60 0 1<br />
16 Cyclopentyl Piperidinyl Methyl Ph(4-Cl) 7.00 7.20 0.20 7.52 7.28 0.24 3.18 0 1<br />
17 Cyclopentyl Piperidinyl Methyl N(Me) 2 6.52 6.15 0.37 6.40 6.07 0.33 0.74 0 1<br />
18 Cyclopentyl Piperidinyl Methyl CF3 6.30 6.64 0.34 6.10 6.63 0.53 1.87 0 1<br />
19 Cyclopentyl Morpholinyl Methyl Methyl 6.40 6.17 0.23 6.10 6.09 0.01 0.77 0 1<br />
20 Cyclopentyl Morpholinyl Methyl Ph(4-OMe) 7.02 6.97 0.05 7.00 7.02 0.02 2.65 0 1<br />
X<br />
O<br />
O<br />
H<br />
N<br />
XV<br />
O<br />
Y<br />
N<br />
H