phylogenetic relationships and classification of didelphid marsupials ...
phylogenetic relationships and classification of didelphid marsupials ...
phylogenetic relationships and classification of didelphid marsupials ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
2009 VOSS AND JANSA: DIDELPHID MARSUPIALS 9<br />
presence <strong>and</strong> absence were both commonly<br />
observed for a given taxon). However,<br />
whereas we formerly (Voss <strong>and</strong> Jansa, 2003)<br />
treated such polymorphism as a separate state<br />
intermediate to the two fixed conditions, with<br />
transformations to <strong>and</strong> from the polymorphic<br />
state weighted as half-steps (the ‘‘scaled’’<br />
option discussed by Wiens, 2000), we here<br />
conform with the prevailing custom <strong>of</strong> coding<br />
polymorphisms as taxonomic ambiguities<br />
(e.g., as ‘‘0/1’’ for a binary character). This<br />
coding change has minimal impact on our<br />
results (only weakly supported <strong>relationships</strong><br />
are affected, even in separate analyses <strong>of</strong><br />
morphology) but it facilitates likelihood<br />
modeling <strong>of</strong> morphological character evolution<br />
(Lewis, 2001) in Bayesian analyses <strong>of</strong> our<br />
combined datasets (see below).<br />
MOLECULAR SEQUENCING AND HOMOLO-<br />
GY COMPARISONS: The laboratory procedures<br />
we used for DNA amplification <strong>and</strong> sequencing<br />
(including the names <strong>and</strong> locations <strong>of</strong><br />
primers used in PCR reactions) have already<br />
been described for three <strong>of</strong> the protein-coding<br />
nuclear loci analyzed in this report: IRBP<br />
(Jansa <strong>and</strong> Voss, 2000), Dentin Matrix<br />
Protein 1 (DMP1; Jansa et al., 2006), <strong>and</strong><br />
Recombination Activating 1 Gene (RAG1;<br />
Gruber et al., 2007). In addition, we<br />
sequenced two other protein-coding nuclear<br />
genes as described below.<br />
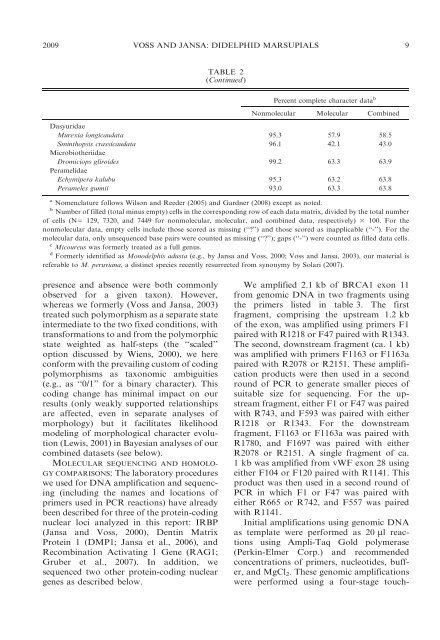
TABLE 2<br />
(Continued)<br />
Percent complete character data b<br />
Nonmolecular Molecular Combined<br />
Dasyuridae<br />
Murexia longicaudata 95.3 57.9 58.5<br />
Sminthopsis crassicaudata<br />
Microbiotheriidae<br />
96.1 42.1 43.0<br />
Dromiciops gliroides<br />
Peramelidae<br />
99.2 63.3 63.9<br />
Echymipera kalubu 95.3 63.2 63.8<br />
Perameles gunnii 93.0 63.3 63.8<br />
a Nomenclature follows Wilson <strong>and</strong> Reeder (2005) <strong>and</strong> Gardner (2008) except as noted.<br />
b Number <strong>of</strong> filled (total minus empty) cells in the corresponding row <strong>of</strong> each data matrix, divided by the total number<br />
<strong>of</strong> cells (N5 129, 7320, <strong>and</strong> 7449 for nonmolecular, molecular, <strong>and</strong> combined data, respectively) 3 100. For the<br />
nonmolecular data, empty cells include those scored as missing (‘‘?’’) <strong>and</strong> those scored as inapplicable (‘‘-’’). For the<br />
molecular data, only unsequenced base pairs were counted as missing (‘‘?’’); gaps (‘‘-’’) were counted as filled data cells.<br />
c Micoureus was formerly treated as a full genus.<br />
d Formerly identified as Monodelphis adusta (e.g., by Jansa <strong>and</strong> Voss, 2000; Voss <strong>and</strong> Jansa, 2003), our material is<br />
referable to M. peruviana, a distinct species recently resurrected from synonymy by Solari (2007).<br />
We amplified 2.1 kb <strong>of</strong> BRCA1 exon 11<br />
from genomic DNA in two fragments using<br />
the primers listed in table 3. The first<br />
fragment, comprising the upstream 1.2 kb<br />
<strong>of</strong> the exon, was amplified using primers F1<br />
paired with R1218 or F47 paired with R1343.<br />
The second, downstream fragment (ca. 1 kb)<br />
was amplified with primers F1163 or F1163a<br />
paired with R2078 or R2151. These amplification<br />
products were then used in a second<br />
round <strong>of</strong> PCR to generate smaller pieces <strong>of</strong><br />
suitable size for sequencing. For the upstream<br />
fragment, either F1 or F47 was paired<br />
with R743, <strong>and</strong> F593 was paired with either<br />
R1218 or R1343. For the downstream<br />
fragment, F1163 or F1163a was paired with<br />
R1780, <strong>and</strong> F1697 was paired with either<br />
R2078 or R2151. A single fragment <strong>of</strong> ca.<br />
1 kb was amplified from vWF exon 28 using<br />
either F104 or F120 paired with R1141. This<br />
product was then used in a second round <strong>of</strong><br />
PCR in which F1 or F47 was paired with<br />
either R665 or R742, <strong>and</strong> F557 was paired<br />
with R1141.<br />
Initial amplifications using genomic DNA<br />
as template were performed as 20 ml reactions<br />
using Ampli-Taq Gold polymerase<br />
(Perkin-Elmer Corp.) <strong>and</strong> recommended<br />
concentrations <strong>of</strong> primers, nucleotides, buffer,<br />
<strong>and</strong> MgCl 2. These genomic amplifications<br />
were performed using a four-stage touch-