- Page 2 and 3:
Computational Biology Editors-in-Ch
- Page 4 and 5:
Kun-Mao Chao·Louxin Zhang Sequence
- Page 6 and 7:
KMC: To Daddy, Mommy, Pei-Pei and L
- Page 8 and 9:
viii Foreword I invite you to study
- Page 10 and 11:
x Preface Chapters 2 to 5 form the
- Page 12 and 13:
Acknowledgments We are extremely gr
- Page 14 and 15:
Contents Foreword .................
- Page 16 and 17:
Contents xix Part II. Theory ......
- Page 18 and 19:
Chapter 1 Introduction 1.1 Biologic
- Page 20 and 21:
1.2 Alignment: A Model for Sequence
- Page 22 and 23:
1.2 Alignment: A Model for Sequence
- Page 24 and 25:
1.3 Scoring Alignment 7 ( ) k m a k
- Page 26 and 27:
1.4 Computing Sequence Alignment 9
- Page 28 and 29:
1.5 Multiple Alignment 11 1.5 Multi
- Page 30 and 31:
1.8 Bibliographic Notes and Further
- Page 32 and 33:
PART I. ALGORITHMS AND TECHNIQUES 1
- Page 34 and 35:
18 2 Basic Algorithmic Techniques 2
- Page 36 and 37:
20 2 Basic Algorithmic Techniques F
- Page 38 and 39:
22 2 Basic Algorithmic Techniques s
- Page 40 and 41:
24 2 Basic Algorithmic Techniques F
- Page 42 and 43: 26 2 Basic Algorithmic Techniques F
- Page 44 and 45: 28 2 Basic Algorithmic Techniques P
- Page 46 and 47: 30 2 Basic Algorithmic Techniques a
- Page 48 and 49: 32 2 Basic Algorithmic Techniques O
- Page 50 and 51: Chapter 3 Pairwise Sequence Alignme
- Page 52 and 53: 3.3 Global Alignment 37 3.2 Dot Mat
- Page 54 and 55: 3.3 Global Alignment 39 ⎧ ⎨ S[i
- Page 56 and 57: 3.3 Global Alignment 41 ( ai b j )
- Page 58 and 59: 3.4 Local Alignment 43 ⎧ 0, ⎪
- Page 60 and 61: 3.4 Local Alignment 45 Algorithm LO
- Page 62 and 63: 3.5 Various Scoring Schemes 47 Fig.
- Page 64 and 65: 3.6 Space-Saving Strategies 49 Fig.
- Page 66 and 67: 3.6 Space-Saving Strategies 51 Algo
- Page 68 and 69: 3.6 Space-Saving Strategies 53 scor
- Page 70 and 71: 3.7 Other Advanced Topics 55 ning,
- Page 72 and 73: 3.7 Other Advanced Topics 57 (0,0).
- Page 74 and 75: 3.7 Other Advanced Topics 59 3.7.4
- Page 76 and 77: 3.8 Bibliographic Notes and Further
- Page 78 and 79: Chapter 4 Homology Search Tools The
- Page 80 and 81: 4.1 Finding Exact Word Matches 65 F
- Page 82 and 83: 4.1 Finding Exact Word Matches 67 F
- Page 84 and 85: 4.3 BLAST 69 SALSDLHAHKLRVDPVNFKLLS
- Page 86 and 87: 4.3 BLAST 71 length w, whereas for
- Page 88 and 89: 4.3 BLAST 73 Fig. 4.9 A scenario of
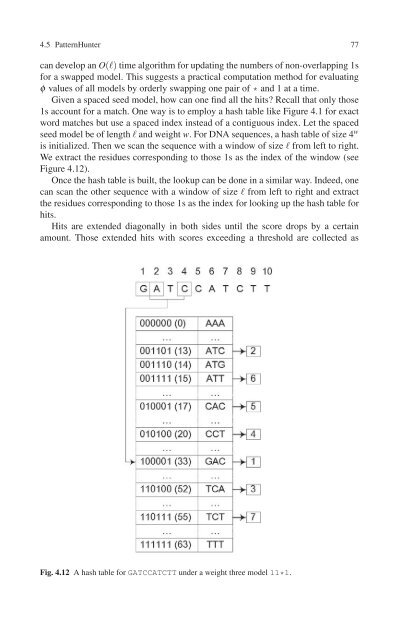
- Page 90 and 91: 4.5 PatternHunter 75 BLAT identifie
- Page 94 and 95: 4.6 Bibliographic Notes and Further
- Page 96 and 97: 82 5 Multiple Sequence Alignment S
- Page 98 and 99: 84 5 Multiple Sequence Alignment S
- Page 100 and 101: 86 5 Multiple Sequence Alignment Fi
- Page 102 and 103: 88 5 Multiple Sequence Alignment al
- Page 104 and 105: Chapter 6 Anatomy of Spaced Seeds B
- Page 106 and 107: 6.2 Basic Formulas on Hit Probabili
- Page 108 and 109: 6.2 Basic Formulas on Hit Probabili
- Page 110 and 111: 6.2 Basic Formulas on Hit Probabili
- Page 112 and 113: 6.3 Distance between Non-Overlappin
- Page 114 and 115: 6.3 Distance between Non-Overlappin
- Page 116 and 117: 6.3 Distance between Non-Overlappin
- Page 118 and 119: 6.4 Asymptotic Analysis of Hit Prob
- Page 120 and 121: 6.4 Asymptotic Analysis of Hit Prob
- Page 122 and 123: 6.4 Asymptotic Analysis of Hit Prob
- Page 124 and 125: 6.5 Spaced Seed Selection 111 Count
- Page 126 and 127: 6.6 Generalizations of Spaced Seeds
- Page 128 and 129: 6.7 Bibliographic Notes and Further
- Page 130 and 131: 6.7 Bibliographic Notes and Further
- Page 132 and 133: 120 7 Local Alignment Statistics tr
- Page 134 and 135: 122 7 Local Alignment Statistics Fi
- Page 136 and 137: 124 7 Local Alignment Statistics Le
- Page 138 and 139: 126 7 Local Alignment Statistics Be
- Page 140 and 141: 128 7 Local Alignment Statistics we
- Page 142 and 143:
130 7 Local Alignment Statistics A
- Page 144 and 145:
132 7 Local Alignment Statistics pr
- Page 146 and 147:
134 7 Local Alignment Statistics wh
- Page 148 and 149:
136 7 Local Alignment Statistics Ta
- Page 150 and 151:
138 7 Local Alignment Statistics Be
- Page 152 and 153:
140 7 Local Alignment Statistics 7.
- Page 154 and 155:
142 7 Local Alignment Statistics 7.
- Page 156 and 157:
144 7 Local Alignment Statistics al
- Page 158 and 159:
146 7 Local Alignment Statistics 7.
- Page 160 and 161:
Chapter 8 Scoring Matrices With the
- Page 162 and 163:
8.1 The PAM Scoring Matrices 151 AB
- Page 164 and 165:
8.2 The BLOSUM Scoring Matrices 153
- Page 166 and 167:
8.3 General Form of the Scoring Mat
- Page 168 and 169:
8.4 How to Select a Scoring Matrix?
- Page 170 and 171:
8.5 Compositional Adjustment of Sco
- Page 172 and 173:
8.6 DNA Scoring Matrices 161 This i
- Page 174 and 175:
8.7 Gap Cost in Gapped Alignments 1
- Page 176 and 177:
8.8 Bibliographic Notes and Further
- Page 178 and 179:
8.8 Bibliographic Notes and Further
- Page 180 and 181:
8.8 Bibliographic Notes and Further
- Page 182 and 183:
8.8 Bibliographic Notes and Further
- Page 184 and 185:
Appendix A Basic Concepts in Molecu
- Page 186 and 187:
A.4 The Genomes 175 ondary structur
- Page 188 and 189:
Appendix B Elementary Probability T
- Page 190 and 191:
B.3 Major Discrete Distributions 17
- Page 192 and 193:
B.3 Major Discrete Distributions 18
- Page 194 and 195:
B.5 Mean, Variance, and Moments 183
- Page 196 and 197:
B.5 Mean, Variance, and Moments 185
- Page 198 and 199:
B.6 Relative Entropy of Probability
- Page 200 and 201:
B.7 Discrete-time Finite Markov Cha
- Page 202 and 203:
B.8 Recurrent Events and the Renewa
- Page 204 and 205:
B.8 Recurrent Events and the Renewa
- Page 206 and 207:
196 C Software Packages for Sequenc
- Page 208 and 209:
198 References 19. Bafna, V. and Pe
- Page 210 and 211:
200 References 71. Fitch, W.M. and
- Page 212 and 213:
202 References 122. Letunic, I., Co
- Page 214 and 215:
204 References 173. Robinson, A.B.
- Page 216 and 217:
Index O-notation, 18 P-value, 139 a
- Page 218:
Index 209 heuristic, 85 progressive