Paterson Institute for Cancer Research SCIENTIFIC REPORT 2005
Paterson Institute for Cancer Research SCIENTIFIC REPORT 2005
Paterson Institute for Cancer Research SCIENTIFIC REPORT 2005
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
54<br />
RESEARCH SERVICES<br />
mised to detect stress activation in mouse embryos<br />
including phosphorylated p38 kinase or hypoxia<br />
specific marker HIF1α.<br />
The unit, together with the Clinical and experimental<br />
pharmacology group, has also been involved in<br />
the optimisation of immunohistochemical (IHC)<br />
methodologies <strong>for</strong> cleaved caspase 3, PARP and<br />
cytokeratin 18 <strong>for</strong> use in pre-clinical pharmacodynamic<br />
IHC assays. The assays focus on detecting<br />
and quantifying levels of apoptosis in cells and tissue<br />
which will eventually be used in the clinical trial<br />
of agents which modulate/inhibit apoptotic pathways.<br />
A new sucrose/gelatin method <strong>for</strong> the preparation<br />
and freezing of embryoid bodies and cell preparations<br />
has been implemented. The method both cryoprotects<br />
and immobilises specimens allowing <strong>for</strong><br />
much improved morphology and numbers.<br />
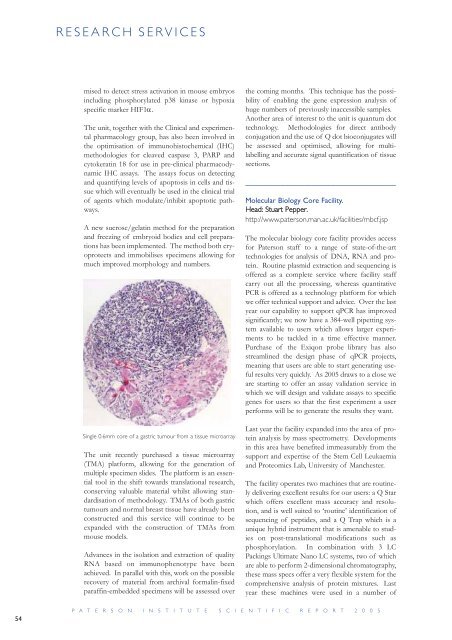
Single 0.6mm core of a gastric tumour from a tissue microarray<br />
The unit recently purchased a tissue microarray<br />
(TMA) plat<strong>for</strong>m, allowing <strong>for</strong> the generation of<br />
multiple specimen slides. The plat<strong>for</strong>m is an essential<br />
tool in the shift towards translational research,<br />
conserving valuable material whilst allowing standardisation<br />
of methodology. TMAs of both gastric<br />
tumours and normal breast tissue have already been<br />
constructed and this service will continue to be<br />
expanded with the construction of TMAs from<br />
mouse models.<br />
Advances in the isolation and extraction of quality<br />
RNA based on immunophenotype have been<br />
achieved. In parallel with this, work on the possible<br />
recovery of material from archival <strong>for</strong>malin-fixed<br />
paraffin-embedded specimens will be assessed over<br />
the coming months. This technique has the possibility<br />
of enabling the gene expression analysis of<br />
huge numbers of previously inaccessible samples.<br />
Another area of interest to the unit is quantum dot<br />
technology. Methodologies <strong>for</strong> direct antibody<br />
conjugation and the use of Q dot bioconjugates will<br />
be assessed and optimised, allowing <strong>for</strong> multilabelling<br />
and accurate signal quantification of tissue<br />
sections.<br />
____________________________________<br />
Molecular Biology Core Facility.<br />
Head: Stuart Pepper.<br />
http://www.paterson.man.ac.uk/facilities/mbcf.jsp<br />
The molecular biology core facility provides access<br />
<strong>for</strong> <strong>Paterson</strong> staff to a range of state-of-the-art<br />
technologies <strong>for</strong> analysis of DNA, RNA and protein.<br />
Routine plasmid extraction and sequencing is<br />
offered as a complete service where facility staff<br />
carry out all the processing, whereas quantitative<br />
PCR is offered as a technology plat<strong>for</strong>m <strong>for</strong> which<br />
we offer technical support and advice. Over the last<br />
year our capability to support qPCR has improved<br />
significantly; we now have a 384-well pipetting system<br />
available to users which allows larger experiments<br />
to be tackled in a time effective manner.<br />
Purchase of the Exiqon probe library has also<br />
streamlined the design phase of qPCR projects,<br />
meaning that users are able to start generating useful<br />
results very quickly. As <strong>2005</strong> draws to a close we<br />
are starting to offer an assay validation service in<br />
which we will design and validate assays to specific<br />
genes <strong>for</strong> users so that the first experiment a user<br />
per<strong>for</strong>ms will be to generate the results they want.<br />
Last year the facility expanded into the area of protein<br />
analysis by mass spectrometry. Developments<br />
in this area have benefited immeasurably from the<br />
support and expertise of the Stem Cell Leukaemia<br />
and Proteomics Lab, University of Manchester.<br />
The facility operates two machines that are routinely<br />
delivering excellent results <strong>for</strong> our users: a Q Star<br />
which offers excellent mass accuracy and resolution,<br />
and is well suited to ‘routine’ identification of<br />
sequencing of peptides, and a Q Trap which is a<br />
unique hybrid instrument that is amenable to studies<br />
on post-translational modifications such as<br />
phosphorylation. In combination with 3 LC<br />
Packings Ultimate Nano LC systems, two of which<br />
are able to per<strong>for</strong>m 2-dimensional chromatography,<br />
these mass specs offer a very flexible system <strong>for</strong> the<br />
comprehensive analysis of protein mixtures. Last<br />
year these machines were used in a number of<br />
P A T E R S O N I N S T I T U T E S C I E N T I F I C R E P O R T 2 0 0 5