Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
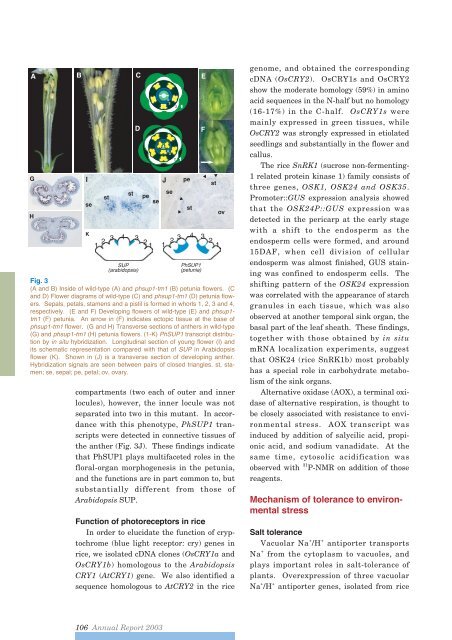
AFig. 3(A and B) Inside of wild-type (A) and phsup1-tm1 (B) petunia flowers. (Cand D) Flower diagrams of wild-type (C) and phsup1-tm1 (D) petunia flowers.Sepals, petals, stamens and a pistil is formed in whorls 1, 2, 3 and 4,respectively. (E and F) Developing flowers of wild-type (E) and phsup1-tm1 (F) petunia. An arrow in (F) indicates ectopic tissue at the base ofphsup1-tm1 flower. (G and H) Transverse sections of anthers in wild-type(G) and phsup1-tm1 (H) petunia flowers. (1-K) PhSUP1 transcript distributionby in situ hybridization. Longitudinal section of young flower (I) andits schematic representation compared with that of SUP in Arabidopsisflower (K). Shown in (J) is a transverse section of developing anther.Hybridization signals are seen between pairs of closed triangles. st, stamen;se, sepal; pe, petal; ov, ovary.compartments (two each of outer and innerlocules), however, the inner locule was notseparated into two in this mutant. In accordancewith this phenotype, PhSUP1 transcriptswere detected in connective tissues ofthe anther (Fig. 3J). These findings indicatethat PhSUP1 plays multifaceted roles in thefloral-organ morphogenesis in the petunia,and the functions are in part common to, butsubstantially different from those ofArabidopsis SUP.Function of photoreceptors in riceIn order to elucidate the function of cryptochrome(blue light receptor: cry) genes inrice, we isolated cDNA clones (OsCRY1a andOsCRY1b) homologous to the ArabidopsisCRY1 (AtCRY1) gene. We also identified asequence homologous to AtCRY2 in the ricegenome, and obtained the correspondingcDNA (OsCRY2). OsCRY1s and OsCRY2show the moderate homology (59%) in aminoacid sequences in the N-half but no homology(16-17%) in the C-half. OsCRY1s weremainly expressed in green tissues, whileOsCRY2 was strongly expressed in etiolatedseedlings and substantially in the flower andcallus.The rice SnRK1 (sucrose non-fermenting-1 related protein kinase 1) family consists ofthree genes, OSK1, OSK24 and OSK35.Promoter::GUS expression analysis showedthat the OSK24P::GUS expression wasdetected in the pericarp at the early stagewith a shift to the endosperm as theendosperm cells were formed, and around15DAF, when cell division of cellularendosperm was almost finished, GUS stainingwas confined to endosperm cells. Theshifting pattern of the OSK24 expressionwas correlated with the appearance of starchgranules in each tissue, which was alsoobserved at another temporal sink organ, thebasal part of the leaf sheath. These findings,together with those obtained by in situmRNA localization experiments, suggestthat OSK24 (rice SnRK1b) most probablyhas a special role in carbohydrate metabolismof the sink organs.Alternative oxidase (AOX), a terminal oxidaseof alternative respiration, is thought tobe closely associated with resistance to environmentalstress. AOX transcript wasinduced by addition of salycilic acid, propionicacid, and sodium vanadidate. At thesame time, cytosolic acidification wasobserved with 31 P-NMR on addition of thosereagents.Mechanism of tolerance to environmentalstressSalt toleranceVacuolar Na + /H +antiporter transportsNa +from the cytoplasm to vacuoles, andplays important roles in salt-tolerance ofplants. Overexpression of three vacuolarNa + /H + antiporter genes, isolated from rice106 <strong>Annual</strong> <strong>Report</strong> <strong>2003</strong>