Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
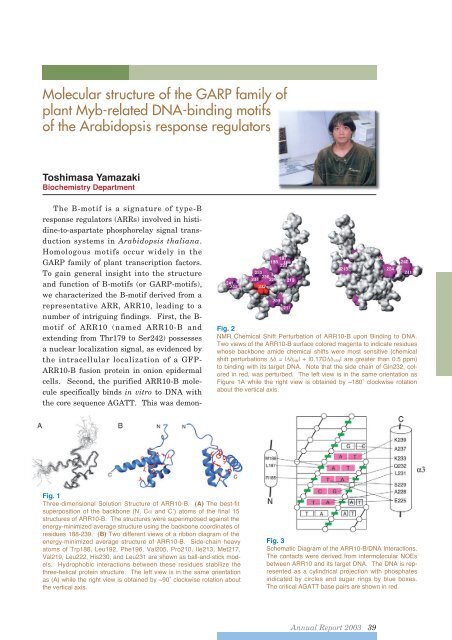
Molecular structure of the GARP family ofplant Myb-related DNA-binding motifsof the Arabidopsis response regulatorsToshimasa YamazakiBiochemistry DepartmentThe B-motif is a signature of type-Bresponse regulators (ARRs) involved in histidine-to-aspartatephosphorelay signal transductionsystems in Arabidopsis thaliana.Homologous motifs occur widely in theGARP family of plant transcription factors.To gain general insight into the structureand function of B-motifs (or GARP-motifs),we characterized the B-motif derived from arepresentative ARR, ARR10, leading to anumber of intriguing findings. First, the B-motif of ARR10 (named ARR10-B andextending from Thr179 to Ser242) possessesa nuclear localization signal, as evidenced bythe intracellular localization of a GFP-ARR10-B fusion protein in onion epidermalcells. Second, the purified ARR10-B moleculespecifically binds in vitro to DNA withthe core sequence AGATT. This was demon-Fig. 2NMR Chemical Shift Perturbation of ARR10-B upon Binding to DNA.Two views of the ARR10-B surface colored magenta to indicate residueswhose backbone amide chemical shifts were most sensitive (chemicalshift perturbations = | HN | + |0.17D 15N | are greater than 0.5 ppm)to binding with its target DNA. Note that the side chain of Gln232, coloredin red, was perturbed. The left view is in the same orientation asFigure 1A while the right view is obtained by ~180˚ clockwise rotationabout the vertical axis.Fig. 1Three-dimensional Solution Structure of ARR10-B. (A) The best-fitsuperposition of the backbone (N, C and C’) atoms of the final 15structures of ARR10-B. The structures were superimposed against theenergy-minimized average structure using the backbone coordinates ofresidues 188-239. (B) Two different views of a ribbon diagram of theenergy-minimized average structure of ARR10-B. Side-chain heavyatoms of Trp188, Leu192, Phe196, Val205, Pro210, Ile213, Met217,Val219, Leu222, His230, and Leu231 are shown as ball-and-stick models.Hydrophobic interactions between these residues stabilize thethree-helical protein structure. The left view is in the same orientationas (A) while the right view is obtained by ~90˚ clockwise rotation aboutthe vertical axis.Fig. 3Schematic Diagram of the ARR10-B/DNA Interactions.The contacts were derived from intermolecular NOEsbetween ARR10 and its target DNA. The DNA is representedas a cylindrical projection with phosphatesindicated by circles and sugar rings by blue boxes.The critical AGATT base pairs are shown in red.<strong>Annual</strong> <strong>Report</strong> <strong>2003</strong> 39