Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
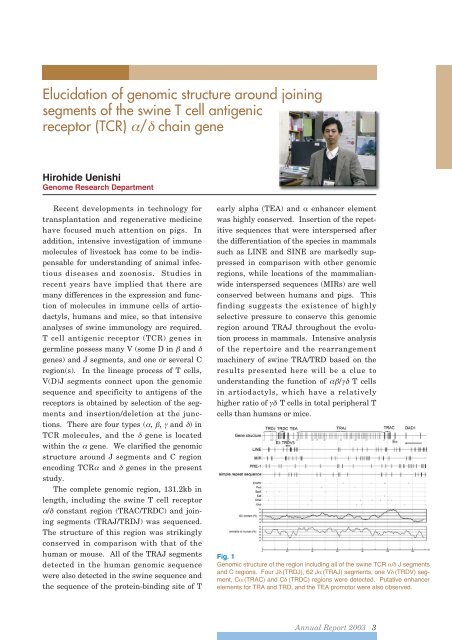
Elucidation of genomic structure around joiningsegments of the swine T cell antigenicreceptor (TCR) / chain geneHirohide UenishiGenome Research DepartmentRecent developments in technology fortransplantation and regenerative medicinehave focused much attention on pigs. Inaddition, intensive investigation of immunemolecules of livestock has come to be indispensablefor understanding of animal infectiousdiseases and zoonosis. Studies inrecent years have implied that there aremany differences in the expression and functionof molecules in immune cells of artiodactyls,humans and mice, so that intensiveanalyses of swine immunology are required.T cell antigenic receptor (TCR) genes ingermline possess many V (some D in and genes) and J segments, and one or several Cregion(s). In the lineage process of T cells,V(D)J segments connect upon the genomicsequence and specificity to antigens of thereceptors is obtained by selection of the segmentsand insertion/deletion at the junctions.There are four types (, , and ) inTCR molecules, and the gene is locatedwithin the gene. We clarified the genomicstructure around J segments and C regionencoding TCR and genes in the presentstudy.The complete genomic region, 131.2kb inlength, including the swine T cell receptor/ constant region (TRAC/TRDC) and joiningsegments (TRAJ/TRDJ) was sequenced.The structure of this region was strikinglyconserved in comparison with that of thehuman or mouse. All of the TRAJ segmentsdetected in the human genomic sequencewere also detected in the swine sequence andthe sequence of the protein-binding site of Tearly alpha (TEA) and enhancer elementwas highly conserved. Insertion of the repetitivesequences that were interspersed afterthe differentiation of the species in mammalssuch as LINE and SINE are markedly suppressedin comparison with other genomicregions, while locations of the mammalianwideinterspersed sequences (MIRs) are wellconserved between humans and pigs. Thisfinding suggests the existence of highlyselective pressure to conserve this genomicregion around TRAJ throughout the evolutionprocess in mammals. Intensive analysisof the repertoire and the rearrangementmachinery of swine TRA/TRD based on theresults presented here will be a clue tounderstanding the function of / T cellsin artiodactyls, which have a relativelyhigher ratio of T cells in total peripheral Tcells than humans or mice.Fig. 1Genomic structure of the region including all of the swine TCR / J segmentsand C regions. Four J (TRDJ), 62 J (TRAJ) segments, one V (TRDV) segment,C (TRAC) and C (TRDC) regions were detected. Putative enhancerelements for TRA and TRD, and the TEA promotor were also observed.<strong>Annual</strong> <strong>Report</strong> <strong>2003</strong> 3