Annual Report 2011 - Center for Advanced Biotechnology and ...
Annual Report 2011 - Center for Advanced Biotechnology and ...
Annual Report 2011 - Center for Advanced Biotechnology and ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
serine 2 residues by pTEF-b, polymerase bound complexes of factor DSIF <strong>and</strong><br />
negative elongation factor (NELF) dissociate <strong>and</strong> processive elongation ensues. In<br />
an ef<strong>for</strong>t to decipher how the critically important human GTase works, we<br />
determined that the minimum enzymatically active domain resides in CE residues<br />
229-567. The<br />
expressed <strong>and</strong> purified active fragment was crystallized <strong>and</strong> the structure<br />
determined by X-ray crystallography. Seven related con<strong>for</strong>mational states were<br />
obtained in the crystal. Position differences of the oligonucleotide/oligosaccharide<br />
(OB) binding fold lid domain over the conserved GTP binding site in the seven<br />
structures provided snapshots of the opening <strong>and</strong> closing of the active site cleft<br />
via a swivel motion. While the GTP binding site is structurally <strong>and</strong> evolutionarily<br />
conserved, the overall GTase mechanism in mammalian <strong>and</strong> yeast systems differs<br />
somewhat. Experiments are underway to crystallize complexes of human GTase<br />
with CTD <strong>and</strong> RNA as well as GTP. Protein engineering is being applied in an<br />
ef<strong>for</strong>t to crystallize the full length human CE.<br />
Publications:<br />
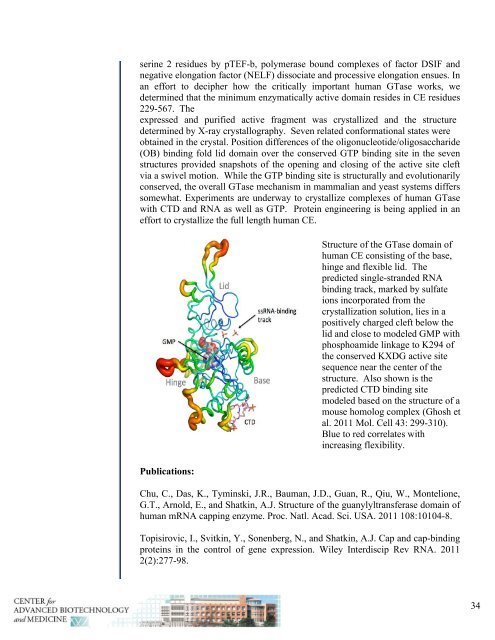
Structure of the GTase domain of<br />
human CE consisting of the base,<br />
hinge <strong>and</strong> flexible lid. The<br />
predicted single-str<strong>and</strong>ed RNA<br />
binding track, marked by sulfate<br />
ions incorporated from the<br />
crystallization solution, lies in a<br />
positively charged cleft below the<br />
lid <strong>and</strong> close to modeled GMP with<br />
phosphoamide linkage to K294 of<br />
the conserved KXDG active site<br />
sequence near the center of the<br />
structure. Also shown is the<br />
predicted CTD binding site<br />
modeled based on the structure of a<br />
mouse homolog complex (Ghosh et<br />
al. <strong>2011</strong> Mol. Cell 43: 299-310).<br />
Blue to red correlates with<br />
increasing flexibility.<br />
Chu, C., Das, K., Tyminski, J.R., Bauman, J.D., Guan, R., Qiu, W., Montelione,<br />
G.T., Arnold, E., <strong>and</strong> Shatkin, A.J. Structure of the guanylyltransferase domain of<br />
human mRNA capping enzyme. Proc. Natl. Acad. Sci. USA. <strong>2011</strong> 108:10104-8.<br />
Topisirovic, I., Svitkin, Y., Sonenberg, N., <strong>and</strong> Shatkin, A.J. Cap <strong>and</strong> cap-binding<br />
proteins in the control of gene expression. Wiley Interdiscip Rev RNA. <strong>2011</strong><br />
2(2):277-98.<br />
34