Casella et al. - 2013 - TILLING in European Rice Hunting Mutations for Cr
Casella et al. - 2013 - TILLING in European Rice Hunting Mutations for Cr
Casella et al. - 2013 - TILLING in European Rice Hunting Mutations for Cr
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
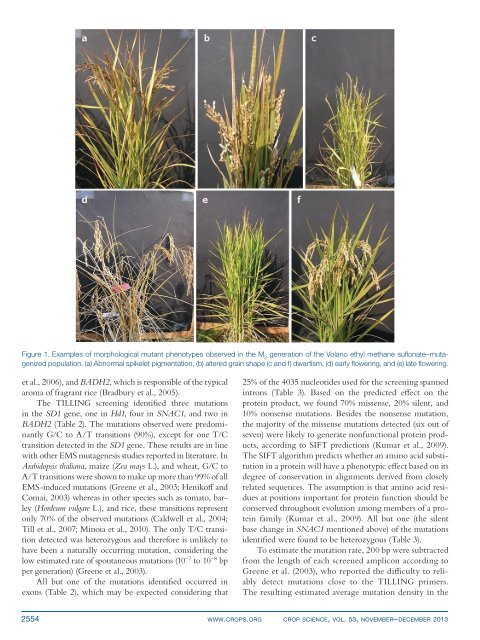
Figure 1. Examples of morphologic<strong>al</strong> mutant phenotypes observed <strong>in</strong> the M 2<br />
generation of the Volano <strong>et</strong>hyl m<strong>et</strong>hane sulfonate–mutagenized<br />
population. (a) Abnorm<strong>al</strong> spikel<strong>et</strong> pigmentation, (b) <strong>al</strong>tered gra<strong>in</strong> shape (c and f) dwarfism, (d) early flower<strong>in</strong>g, and (e) late flower<strong>in</strong>g.<br />
<strong>et</strong> <strong>al</strong>., 2006), and BADH2, which is responsible of the typic<strong>al</strong><br />
aroma of fragrant rice (Bradbury <strong>et</strong> <strong>al</strong>., 2005).<br />
The <strong>TILLING</strong> screen<strong>in</strong>g identified three mutations<br />
<strong>in</strong> the SD1 gene, one <strong>in</strong> Hd1, four <strong>in</strong> SNAC1, and two <strong>in</strong><br />
BADH2 (Table 2). The mutations observed were predom<strong>in</strong>antly<br />
G/C to A/T transitions (90%), except <strong>for</strong> one T/C<br />
transition d<strong>et</strong>ected <strong>in</strong> the SD1 gene. These results are <strong>in</strong> l<strong>in</strong>e<br />
with other EMS mutagenesis studies reported <strong>in</strong> literature. In<br />
Arabidopsis th<strong>al</strong>iana, maize (Zea mays L.), and wheat, G/C to<br />
A/T transitions were shown to make up more than 99% of <strong>al</strong>l<br />
EMS-<strong>in</strong>duced mutations (Greene <strong>et</strong> <strong>al</strong>., 2003; Henikoff and<br />
Comai, 2003) whereas <strong>in</strong> other species such as tomato, barley<br />
(Hordeum vulgare L.), and rice, these transitions represent<br />
only 70% of the observed mutations (C<strong>al</strong>dwell <strong>et</strong> <strong>al</strong>., 2004;<br />
Till <strong>et</strong> <strong>al</strong>., 2007; M<strong>in</strong>oia <strong>et</strong> <strong>al</strong>., 2010). The only T/C transition<br />
d<strong>et</strong>ected was h<strong>et</strong>erozygous and there<strong>for</strong>e is unlikely to<br />
have been a natur<strong>al</strong>ly occurr<strong>in</strong>g mutation, consider<strong>in</strong>g the<br />
low estimated rate of spontaneous mutations (10 –7 to 10 –8 bp<br />
per generation) (Greene <strong>et</strong> <strong>al</strong>., 2003).<br />
All but one of the mutations identified occurred <strong>in</strong><br />
exons (Table 2), which may be expected consider<strong>in</strong>g that<br />
25% of the 4035 nucleotides used <strong>for</strong> the screen<strong>in</strong>g spanned<br />
<strong>in</strong>trons (Table 3). Based on the predicted effect on the<br />
prote<strong>in</strong> product, we found 70% missense, 20% silent, and<br />
10% nonsense mutations. Besides the nonsense mutation,<br />
the majority of the missense mutations d<strong>et</strong>ected (six out of<br />
seven) were likely to generate nonfunction<strong>al</strong> prote<strong>in</strong> products,<br />
accord<strong>in</strong>g to SIFT predictions (Kumar <strong>et</strong> <strong>al</strong>., 2009).<br />
The SIFT <strong>al</strong>gorithm predicts wh<strong>et</strong>her an am<strong>in</strong>o acid substitution<br />
<strong>in</strong> a prote<strong>in</strong> will have a phenotypic effect based on its<br />
degree of conservation <strong>in</strong> <strong>al</strong>ignments derived from closely<br />
related sequences. The assumption is that am<strong>in</strong>o acid residues<br />
at positions important <strong>for</strong> prote<strong>in</strong> function should be<br />
conserved throughout evolution among members of a prote<strong>in</strong><br />
family (Kumar <strong>et</strong> <strong>al</strong>., 2009). All but one (the silent<br />
base change <strong>in</strong> SNAC1 mentioned above) of the mutations<br />
identified were found to be h<strong>et</strong>erozygous (Table 3).<br />
To estimate the mutation rate, 200 bp were subtracted<br />
from the length of each screened amplicon accord<strong>in</strong>g to<br />
Greene <strong>et</strong> <strong>al</strong>. (2003), who reported the difficulty to reliably<br />
d<strong>et</strong>ect mutations close to the <strong>TILLING</strong> primers.<br />
The result<strong>in</strong>g estimated average mutation density <strong>in</strong> the<br />
2554 www.crops.org crop science, vol. 53, november–december <strong>2013</strong>