New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
4.2. IMPROVED KERNELS FOR MHC BINDING PREDICTION 37<br />
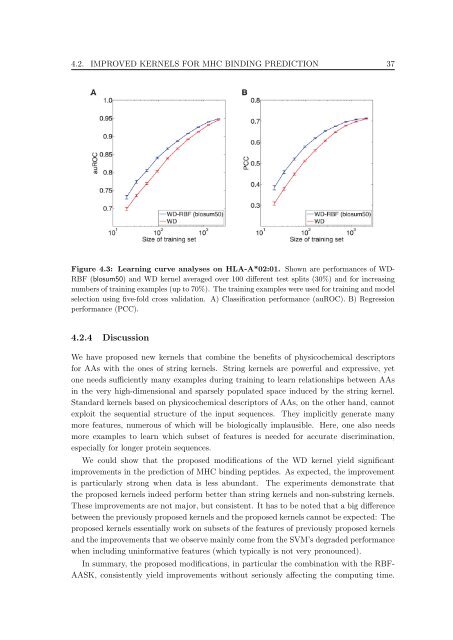
Figure 4.3: Learn<strong>in</strong>g curve analyses on HLA-A*02:01. Shown are performances <strong>of</strong> WD-<br />
RBF (blosum50) and WD kernel averaged over 100 different test splits (30%) and for <strong>in</strong>creas<strong>in</strong>g<br />
numbers <strong>of</strong> tra<strong>in</strong><strong>in</strong>g examples (up <strong>to</strong> 70%). The tra<strong>in</strong><strong>in</strong>g examples were used for tra<strong>in</strong><strong>in</strong>g and model<br />
selection us<strong>in</strong>g five-fold cross validation. A) Classification performance (auROC). B) Regression<br />
performance (PCC).<br />
4.2.4 Discussion<br />
We have proposed new kernels that comb<strong>in</strong>e the benefits <strong>of</strong> physicochemical descrip<strong>to</strong>rs<br />
for AAs with the ones <strong>of</strong> str<strong>in</strong>g kernels. Str<strong>in</strong>g kernels are powerful and expressive, yet<br />
one needs sufficiently many examples dur<strong>in</strong>g tra<strong>in</strong><strong>in</strong>g <strong>to</strong> learn relationships between AAs<br />
<strong>in</strong> the very high-dimensional and sparsely populated space <strong>in</strong>duced by the str<strong>in</strong>g kernel.<br />
Standard kernels based on physicochemical descrip<strong>to</strong>rs <strong>of</strong> AAs, on the other hand, cannot<br />
exploit the sequential structure <strong>of</strong> the <strong>in</strong>put sequences. They implicitly generate many<br />
more features, numerous <strong>of</strong> which will be biologically implausible. Here, one also needs<br />
more examples <strong>to</strong> learn which subset <strong>of</strong> features is needed for accurate discrim<strong>in</strong>ation,<br />
especially for longer prote<strong>in</strong> sequences.<br />
We could show that the proposed modifications <strong>of</strong> the WD kernel yield significant<br />
improvements <strong>in</strong> the prediction <strong>of</strong> MHC b<strong>in</strong>d<strong>in</strong>g peptides. As expected, the improvement<br />
is particularly strong when data is less abundant. The experiments demonstrate that<br />
the proposed kernels <strong>in</strong>deed perform better than str<strong>in</strong>g kernels and non-substr<strong>in</strong>g kernels.<br />
These improvements are not major, but consistent. It has <strong>to</strong> be noted that a big difference<br />
between the previously proposed kernels and the proposed kernels cannot be expected: The<br />
proposed kernels essentially work on subsets <strong>of</strong> the features <strong>of</strong> previously proposed kernels<br />
and the improvements that we observe ma<strong>in</strong>ly come from the SVM’s degraded performance<br />
when <strong>in</strong>clud<strong>in</strong>g un<strong>in</strong>formative features (which typically is not very pronounced).<br />
In summary, the proposed modifications, <strong>in</strong> particular the comb<strong>in</strong>ation with the RBF-<br />
AASK, consistently yield improvements without seriously affect<strong>in</strong>g the comput<strong>in</strong>g time.