New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
4.4. T-CELL EPITOPE PREDICTION 49<br />
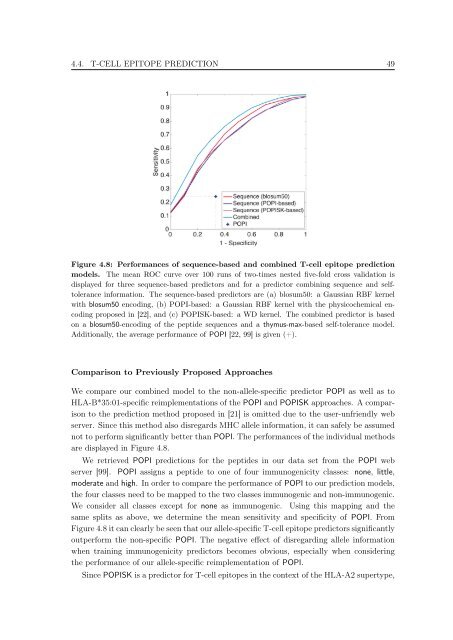
Figure 4.8: Performances <strong>of</strong> sequence-based and comb<strong>in</strong>ed T-cell epi<strong>to</strong>pe prediction<br />
models. The mean ROC curve over 100 runs <strong>of</strong> two-times nested five-fold cross validation is<br />
displayed for three sequence-based predic<strong>to</strong>rs and for a predic<strong>to</strong>r comb<strong>in</strong><strong>in</strong>g sequence and self<strong>to</strong>lerance<br />
<strong>in</strong>formation. The sequence-based predic<strong>to</strong>rs are (a) blosum50: a Gaussian RBF kernel<br />
with blosum50 encod<strong>in</strong>g, (b) POPI-based: a Gaussian RBF kernel with the physicochemical encod<strong>in</strong>g<br />
proposed <strong>in</strong> [22], and (c) POPISK-based: a WD kernel. The comb<strong>in</strong>ed predic<strong>to</strong>r is based<br />
on a blosum50-encod<strong>in</strong>g <strong>of</strong> the peptide sequences and a thymus-max-based self-<strong>to</strong>lerance model.<br />
Additionally, the average performance <strong>of</strong> POPI [22, 99] is given (+).<br />
Comparison <strong>to</strong> Previously Proposed <strong>Approaches</strong><br />
We compare our comb<strong>in</strong>ed model <strong>to</strong> the non-allele-specific predic<strong>to</strong>r POPI as well as <strong>to</strong><br />
HLA-B*35:01-specific reimplementations <strong>of</strong> the POPI and POPISK approaches. A comparison<br />
<strong>to</strong> the prediction method proposed <strong>in</strong> [21] is omitted due <strong>to</strong> the user-unfriendly web<br />
server. S<strong>in</strong>ce this method also disregards MHC allele <strong>in</strong>formation, it can safely be assumed<br />
not <strong>to</strong> perform significantly better than POPI. The performances <strong>of</strong> the <strong>in</strong>dividual methods<br />
are displayed <strong>in</strong> Figure 4.8.<br />
We retrieved POPI predictions for the peptides <strong>in</strong> our data set from the POPI web<br />
server [99]. POPI assigns a peptide <strong>to</strong> one <strong>of</strong> four immunogenicity classes: none, little,<br />
moderate and high. In order <strong>to</strong> compare the performance <strong>of</strong> POPI <strong>to</strong> our prediction models,<br />
the four classes need <strong>to</strong> be mapped <strong>to</strong> the two classes immunogenic and non-immunogenic.<br />
We consider all classes except for none as immunogenic. Us<strong>in</strong>g this mapp<strong>in</strong>g and the<br />
same splits as above, we determ<strong>in</strong>e the mean sensitivity and specificity <strong>of</strong> POPI. From<br />
Figure 4.8 it can clearly be seen that our allele-specific T-cell epi<strong>to</strong>pe predic<strong>to</strong>rs significantly<br />
outperform the non-specific POPI. The negative effect <strong>of</strong> disregard<strong>in</strong>g allele <strong>in</strong>formation<br />
when tra<strong>in</strong><strong>in</strong>g immunogenicity predic<strong>to</strong>rs becomes obvious, especially when consider<strong>in</strong>g<br />
the performance <strong>of</strong> our allele-specific reimplementation <strong>of</strong> POPI.<br />
S<strong>in</strong>ce POPISK is a predic<strong>to</strong>r for T-cell epi<strong>to</strong>pes <strong>in</strong> the context <strong>of</strong> the HLA-A2 supertype,