New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
6.4. EXPERIMENTAL RESULTS 69<br />
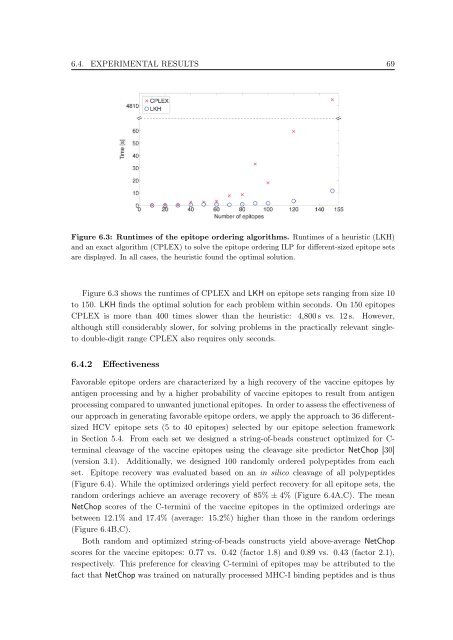
Figure 6.3: Runtimes <strong>of</strong> the epi<strong>to</strong>pe order<strong>in</strong>g algorithms. Runtimes <strong>of</strong> a heuristic (LKH)<br />
and an exact algorithm (CPLEX) <strong>to</strong> solve the epi<strong>to</strong>pe order<strong>in</strong>g ILP for different-sized epi<strong>to</strong>pe sets<br />
are displayed. In all cases, the heuristic found the optimal solution.<br />
Figure 6.3 shows the runtimes <strong>of</strong> CPLEX and LKH on epi<strong>to</strong>pe sets rang<strong>in</strong>g from size 10<br />
<strong>to</strong> 150. LKH f<strong>in</strong>ds the optimal solution for each problem with<strong>in</strong> seconds. On 150 epi<strong>to</strong>pes<br />
CPLEX is more than 400 times slower than the heuristic: 4,800 s vs. 12 s. However,<br />
although still considerably slower, for solv<strong>in</strong>g problems <strong>in</strong> the practically relevant s<strong>in</strong>gle<strong>to</strong><br />
double-digit range CPLEX also requires only seconds.<br />
6.4.2 Effectiveness<br />
Favorable epi<strong>to</strong>pe orders are characterized by a high recovery <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes by<br />
antigen process<strong>in</strong>g and by a higher probability <strong>of</strong> vacc<strong>in</strong>e epi<strong>to</strong>pes <strong>to</strong> result from antigen<br />
process<strong>in</strong>g compared <strong>to</strong> unwanted junctional epi<strong>to</strong>pes. In order <strong>to</strong> assess the effectiveness <strong>of</strong><br />
our approach <strong>in</strong> generat<strong>in</strong>g favorable epi<strong>to</strong>pe orders, we apply the approach <strong>to</strong> 36 differentsized<br />
HCV epi<strong>to</strong>pe sets (5 <strong>to</strong> 40 epi<strong>to</strong>pes) selected by our epi<strong>to</strong>pe selection framework<br />
<strong>in</strong> Section 5.4. From each set we designed a str<strong>in</strong>g-<strong>of</strong>-beads construct optimized for Cterm<strong>in</strong>al<br />
cleavage <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes us<strong>in</strong>g the cleavage site predic<strong>to</strong>r NetChop [30]<br />
(version 3.1). Additionally, we designed 100 randomly ordered polypeptides from each<br />
set. Epi<strong>to</strong>pe recovery was evaluated based on an <strong>in</strong> <strong>silico</strong> cleavage <strong>of</strong> all polypeptides<br />
(Figure 6.4). While the optimized order<strong>in</strong>gs yield perfect recovery for all epi<strong>to</strong>pe sets, the<br />
random order<strong>in</strong>gs achieve an average recovery <strong>of</strong> 85% ± 4% (Figure 6.4A,C). The mean<br />
NetChop scores <strong>of</strong> the C-term<strong>in</strong>i <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes <strong>in</strong> the optimized order<strong>in</strong>gs are<br />
between 12.1% and 17.4% (average: 15.2%) higher than those <strong>in</strong> the random order<strong>in</strong>gs<br />
(Figure 6.4B,C).<br />
Both random and optimized str<strong>in</strong>g-<strong>of</strong>-beads constructs yield above-average NetChop<br />
scores for the vacc<strong>in</strong>e epi<strong>to</strong>pes: 0.77 vs. 0.42 (fac<strong>to</strong>r 1.8) and 0.89 vs. 0.43 (fac<strong>to</strong>r 2.1),<br />
respectively. This preference for cleav<strong>in</strong>g C-term<strong>in</strong>i <strong>of</strong> epi<strong>to</strong>pes may be attributed <strong>to</strong> the<br />
fact that NetChop was tra<strong>in</strong>ed on naturally processed MHC-I b<strong>in</strong>d<strong>in</strong>g peptides and is thus