New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
7.2. DESIGN OF A PEPTIDE COCKTAIL VACCINE 81<br />
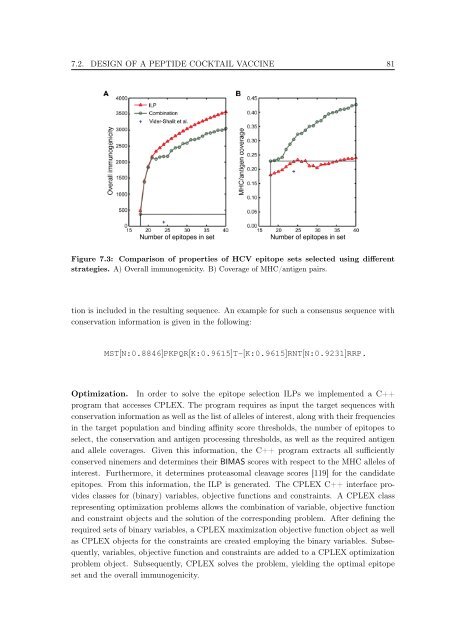
Number <strong>of</strong> epi<strong>to</strong>pes <strong>in</strong> set Number <strong>of</strong> epi<strong>to</strong>pes <strong>in</strong> set<br />
Figure 7.3: Comparison <strong>of</strong> properties <strong>of</strong> HCV epi<strong>to</strong>pe sets selected us<strong>in</strong>g different<br />
strategies. A) Overall immunogenicity. B) Coverage <strong>of</strong> MHC/antigen pairs.<br />
tion is <strong>in</strong>cluded <strong>in</strong> the result<strong>in</strong>g sequence. An example for such a consensus sequence with<br />
conservation <strong>in</strong>formation is given <strong>in</strong> the follow<strong>in</strong>g:<br />
MST[N:0.8846]PKPQR[K:0.9615]T-[K:0.9615]RNT[N:0.9231]RRP.<br />
Optimization. In order <strong>to</strong> solve the epi<strong>to</strong>pe selection ILPs we implemented a C++<br />
program that accesses CPLEX. The program requires as <strong>in</strong>put the target sequences with<br />
conservation <strong>in</strong>formation as well as the list <strong>of</strong> alleles <strong>of</strong> <strong>in</strong>terest, along with their frequencies<br />
<strong>in</strong> the target population and b<strong>in</strong>d<strong>in</strong>g aff<strong>in</strong>ity score thresholds, the number <strong>of</strong> epi<strong>to</strong>pes <strong>to</strong><br />
select, the conservation and antigen process<strong>in</strong>g thresholds, as well as the required antigen<br />
and allele coverages. Given this <strong>in</strong>formation, the C++ program extracts all sufficiently<br />
conserved n<strong>in</strong>emers and determ<strong>in</strong>es their BIMAS scores with respect <strong>to</strong> the MHC alleles <strong>of</strong><br />
<strong>in</strong>terest. Furthermore, it determ<strong>in</strong>es proteasomal cleavage scores [119] for the candidate<br />
epi<strong>to</strong>pes. From this <strong>in</strong>formation, the ILP is generated. The CPLEX C++ <strong>in</strong>terface provides<br />
classes for (b<strong>in</strong>ary) variables, objective functions and constra<strong>in</strong>ts. A CPLEX class<br />
represent<strong>in</strong>g optimization problems allows the comb<strong>in</strong>ation <strong>of</strong> variable, objective function<br />
and constra<strong>in</strong>t objects and the solution <strong>of</strong> the correspond<strong>in</strong>g problem. After def<strong>in</strong><strong>in</strong>g the<br />
required sets <strong>of</strong> b<strong>in</strong>ary variables, a CPLEX maximization objective function object as well<br />
as CPLEX objects for the constra<strong>in</strong>ts are created employ<strong>in</strong>g the b<strong>in</strong>ary variables. Subsequently,<br />
variables, objective function and constra<strong>in</strong>ts are added <strong>to</strong> a CPLEX optimization<br />
problem object. Subsequently, CPLEX solves the problem, yield<strong>in</strong>g the optimal epi<strong>to</strong>pe<br />
set and the overall immunogenicity.