New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
New Approaches to in silico Design of Epitope-Based Vaccines
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
70 CHAPTER 6. EPITOPE ASSEMBLY<br />
A B<br />
C<br />
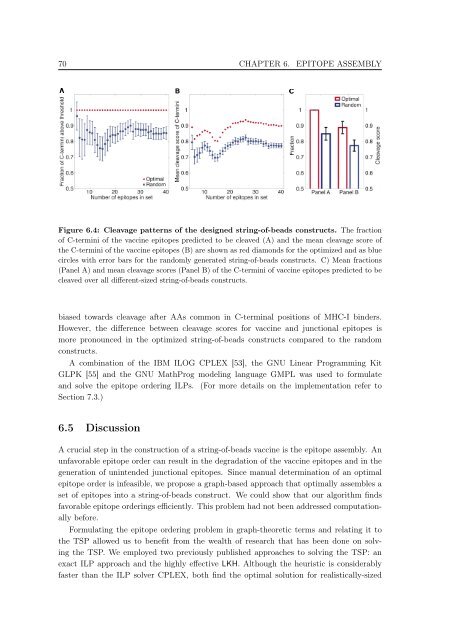
Figure 6.4: Cleavage patterns <strong>of</strong> the designed str<strong>in</strong>g-<strong>of</strong>-beads constructs. The fraction<br />
<strong>of</strong> C-term<strong>in</strong>i <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes predicted <strong>to</strong> be cleaved (A) and the mean cleavage score <strong>of</strong><br />
the C-term<strong>in</strong>i <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes (B) are shown as red diamonds for the optimized and as blue<br />
circles with error bars for the randomly generated str<strong>in</strong>g-<strong>of</strong>-beads constructs. C) Mean fractions<br />
(Panel A) and mean cleavage scores (Panel B) <strong>of</strong> the C-term<strong>in</strong>i <strong>of</strong> vacc<strong>in</strong>e epi<strong>to</strong>pes predicted <strong>to</strong> be<br />
cleaved over all different-sized str<strong>in</strong>g-<strong>of</strong>-beads constructs.<br />
biased <strong>to</strong>wards cleavage after AAs common <strong>in</strong> C-term<strong>in</strong>al positions <strong>of</strong> MHC-I b<strong>in</strong>ders.<br />
However, the difference between cleavage scores for vacc<strong>in</strong>e and junctional epi<strong>to</strong>pes is<br />
more pronounced <strong>in</strong> the optimized str<strong>in</strong>g-<strong>of</strong>-beads constructs compared <strong>to</strong> the random<br />
constructs.<br />
A comb<strong>in</strong>ation <strong>of</strong> the IBM ILOG CPLEX [53], the GNU L<strong>in</strong>ear Programm<strong>in</strong>g Kit<br />
GLPK [55] and the GNU MathProg model<strong>in</strong>g language GMPL was used <strong>to</strong> formulate<br />
and solve the epi<strong>to</strong>pe order<strong>in</strong>g ILPs. (For more details on the implementation refer <strong>to</strong><br />
Section 7.3.)<br />
6.5 Discussion<br />
A crucial step <strong>in</strong> the construction <strong>of</strong> a str<strong>in</strong>g-<strong>of</strong>-beads vacc<strong>in</strong>e is the epi<strong>to</strong>pe assembly. An<br />
unfavorable epi<strong>to</strong>pe order can result <strong>in</strong> the degradation <strong>of</strong> the vacc<strong>in</strong>e epi<strong>to</strong>pes and <strong>in</strong> the<br />
generation <strong>of</strong> un<strong>in</strong>tended junctional epi<strong>to</strong>pes. S<strong>in</strong>ce manual determ<strong>in</strong>ation <strong>of</strong> an optimal<br />
epi<strong>to</strong>pe order is <strong>in</strong>feasible, we propose a graph-based approach that optimally assembles a<br />
set <strong>of</strong> epi<strong>to</strong>pes <strong>in</strong><strong>to</strong> a str<strong>in</strong>g-<strong>of</strong>-beads construct. We could show that our algorithm f<strong>in</strong>ds<br />
favorable epi<strong>to</strong>pe order<strong>in</strong>gs efficiently. This problem had not been addressed computationally<br />
before.<br />
Formulat<strong>in</strong>g the epi<strong>to</strong>pe order<strong>in</strong>g problem <strong>in</strong> graph-theoretic terms and relat<strong>in</strong>g it <strong>to</strong><br />
the TSP allowed us <strong>to</strong> benefit from the wealth <strong>of</strong> research that has been done on solv<strong>in</strong>g<br />
the TSP. We employed two previously published approaches <strong>to</strong> solv<strong>in</strong>g the TSP: an<br />
exact ILP approach and the highly effective LKH. Although the heuristic is considerably<br />
faster than the ILP solver CPLEX, both f<strong>in</strong>d the optimal solution for realistically-sized