You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>EMBL</strong> Hamburg<br />
Macromolecular crystallography<br />
Accurate modelling of biological processes depends on an intimate knowledge of the macromolecules<br />
involved. The availability of models of the interacting components allows a deep understanding<br />
of the basic processes that underlie human disease and all other cellular events. One<br />
recent example is a basis for neurodegenerative pathologies provided by the structure of a stable<br />
serpin dimer (Yamasaki et al., 2008, Nature).<br />
Macromolecular structures are produced by a large number of independent scientists, all of whom<br />
approach specific problems in their own manner. Methods are needed to establish objective structure<br />
determination and quality assessment, which will impact further system-wide analysis. The<br />
group’s major activity is development of novel approaches and software tools for high-throughput<br />
3D structure determination and detailed interpretation of biological macromolecules and their<br />
complexes. Specific attention is given to extension of the current limits of applicability of MX and<br />
its use with complementary techniques.<br />
Previous and current research<br />
In MX, availability of comprehensive software packages has a major impact on structural biology.<br />
Traditionally crystallographic model building is done by expert users with the aid of specialised<br />
graphics software. The automation of this process, first exemplified in the ARP/wARP package<br />
(Langer et al., 2008, Nature Protocols), was promptly followed by developments worldwide.<br />
ARP/wARP has been used extensively for thousands of structure determinations and is often used<br />
as a benchmark to evaluate the quality of electron density obtained by new methods; it has been integrated into many crystallographic software<br />
pipelines as the default model-building engine. A wide spectrum of ARP/wARP functionalities (e.g. Hattne et al., 2008, Acta Cryst.) makes<br />
the software particularly attractive to users.<br />
We continue the developments of the automated structure determination pipeline AutoRickshaw (led by S. Panjikar in collaboration with the<br />
Tucker group and Weiss team (pages 106 and 107)), which uses an artificial intelligence decision-making system (Manjasetty et al., 2008,<br />
Proteomics). With the help of ARP/wARP and other software, AutoRickshaw produces<br />
an interpretable electron density map and a partial structure shortly after data<br />
acquisition. At our MX beamlines the users receive immediate feedback whether the<br />
measured data are of sufficient quality for successful structure determination.<br />
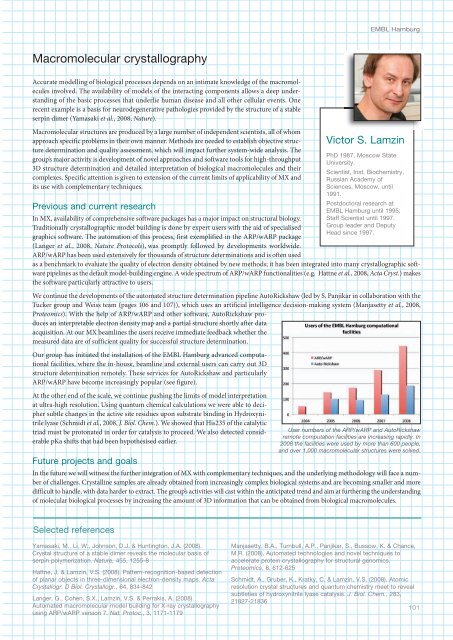
Our group has initiated the installation of the <strong>EMBL</strong> Hamburg advanced computational<br />
facilities, where the in-house, beamline and external users can carry out 3D<br />
structure determination remotely. These services for AutoRickshaw and particularly<br />
ARP/wARP have become increasingly popular (see figure).<br />
At the other end of the scale, we continue pushing the limits of model interpretation<br />
at ultra-high resolution. Using quantum chemical calculations we were able to decipher<br />
subtle changes in the active site residues upon substrate binding in Hydroxynitrile<br />
lyase (Schmidt et al., 2008, J. Biol. Chem.). We showed that His235 of the catalytic<br />
triad must be protonated in order for catalysis to proceed. We also detected considerable<br />
pKa shifts that had been hypothesised earlier.<br />
Future projects and goals<br />
Victor S. Lamzin<br />
PhD 1987, Moscow State<br />
University.<br />
Scientist, Inst. Biochemistry,<br />
Russian Academy of<br />
Sciences, Moscow, until<br />
1991.<br />
Postdoctoral research at<br />
<strong>EMBL</strong> Hamburg until 1995;<br />
Staff Scientist until 1997.<br />
Group leader and Deputy<br />
Head since 1997.<br />
User numbers of the ARP/wARP and AutoRickshaw<br />
remote computation facilities are increasing rapidly. In<br />
2008 the facilities were used by more than 600 people,<br />
and over 1,000 macromolecular structures were solved.<br />
In the future we will witness the further integration of MX with complementary techniques, and the underlying methodology will face a number<br />
of challenges. Crystalline samples are already obtained from increasingly complex biological systems and are becoming smaller and more<br />
difficult to handle, with data harder to extract. The group’s activities will cast within the anticipated trend and aim at furthering the understanding<br />
of molecular biological processes by increasing the amount of 3D information that can be obtained from biological macromolecules.<br />
Selected references<br />
Yamasaki, M., Li, W., Johnson, D.J. & Huntington, J.A. (2008).<br />
Crystal structure of a stable dimer reveals the molecular basis of<br />
serpin polymerization. Nature, 55, 1255-8<br />
Hattne, J. & Lamzin, V.S. (2008). Pattern-recognition-based detection<br />
of planar objects in three-dimensional electron-density maps. Acta<br />
Crystallogr. D Biol. Crystallogr., 6, 83-82<br />
Langer, G., Cohen, S.X., Lamzin, V.S. & Perrakis, A. (2008).<br />
Automated macromolecular model building for X-ray crystallography<br />
using ARP/wARP version 7. Nat. Protoc., 3, 1171-1179<br />
Manjasetty, B.A., Turnbull, A.P., Panjikar, S., Bussow, K. & Chance,<br />
M.R. (2008). Automated technologies and novel techniques to<br />
accelerate protein crystallography for structural genomics.<br />
Proteomics, 8, 612-625<br />
Schmidt, A., Gruber, K., Kratky, C. & Lamzin, V.S. (2008). Atomic<br />
resolution crystal structures and quantum chemistry meet to reveal<br />
subtleties of hydroxynitrile lyase catalysis. J. Biol. Chem., 283,<br />
21827-21836<br />
101