Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>EMBL</strong> Research at a Glance 2009<br />
Jan Ellenberg<br />
PhD 1998, Freie Universität<br />
Berlin.<br />
Postdoctoral research at the<br />
Cell Biology and Metabolism<br />
Branch, NICHD, NIH,<br />
Bethesda.<br />
Group leader at <strong>EMBL</strong> since<br />
1999.<br />
Head of Gene Expression<br />
Unit since 2006. Joint Unit<br />
Coordinator of Cell Biology<br />
and Biophysics Unit since<br />
2009.<br />
Functional dynamics of nuclear structure during<br />
the cell cycle<br />
Previous and current research<br />
The genome of eukaryotic cells is compartmentalised inside the nucleus, delimited by the nuclear<br />
envelope (NE) whose double membranes are continuous with the endoplasmatic reticulum (ER)<br />
and stabilised by the nuclear lamina filament meshwork. The NE is perforated by nuclear pore<br />
complexes (NPCs), which allow selective traffic between nucleus and cytoplasm. In M-phase, most<br />
metazoan cells reversibly dismantle the highly ordered structure of the NE. Nuclear membranes<br />
that surround chromatin in interphase are ‘replaced’ by cytoplasmic spindle microtubules, which<br />
segregate the condensed chromosomes in an ‘open’ division. After chromosome segregation the<br />
nucleus rapidly reassembles.<br />
The overall aim of our research is to elucidate the mechanisms underlying cell cycle remodelling<br />
of the nucleus in live cells. Breakdown and reassembly of the nucleus and the formation and correct<br />
movement of compact mitotic chromosomes are essential but poorly understood processes.<br />
To study them, we are assaying fluorescently-tagged structural proteins and their regulators using<br />
advanced fluorescence microscopy methods coupled with computerised image processing and<br />
simulations to extract biophysical parameters and build mechanistic models.<br />
In the past, we could define<br />
the ER as the reservoir and<br />
means of partitioning for<br />
nuclear membrane proteins<br />
in mitosis and found that nuclear breakdown is triggered by disassembly<br />
of the NPC and then further facilitated by microtubule mediated<br />
tearing of the nuclear lamina. During the meiotic division of starfish<br />
oocytes, we could show that long-range chromosome motion after nuclear<br />
breakdown is driven by actin filaments. In mouse oocytes this occurs<br />
only after formation of an acentrosomal spindle, which we could<br />
show assembles by self-organisation of cytoplasmic microtubule asters.<br />
In mitotic cells, we have analysed chromosome dynamics during their<br />
segregation and could show that their overall spatial arrangement is<br />
transmitted through mitosis and that their maximal compaction is<br />
reached only at the end of anaphase, just before nuclear reformation.<br />
Future projects and goals<br />
The objective of our future work is to gain further mechanistic insight<br />
into nuclear remodelling in live cells. In particular, we are focussing on<br />
the mechanism of nuclear growth in interphase, nuclear disassembly<br />
and reformation as well as chromosome condensation and positioning<br />
in somatic cells and microtubule-independent chromosome motion in<br />
oocytes. To rapidly obtain quantitative data from intact cells, we aim to<br />
automate and standardise advanced fluorescence microscopy assays as<br />
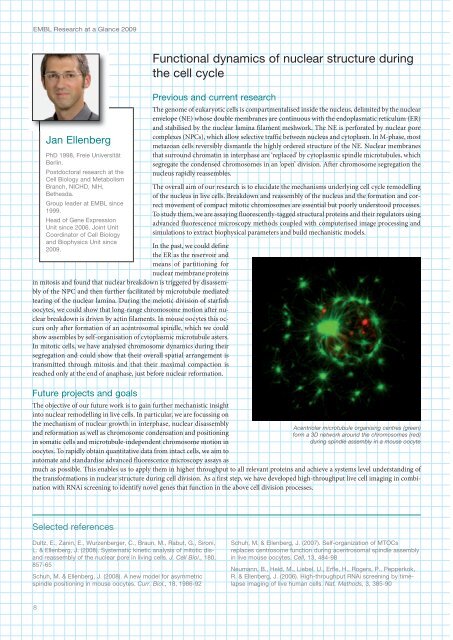
Acentriolar microtubule organising centres (green)<br />
form a 3D network around the chromosomes (red)<br />
during spindle assembly in a mouse oocyte<br />
much as possible. This enables us to apply them in higher throughput to all relevant proteins and achieve a systems level understanding of<br />
the transformations in nuclear structure during cell division. As a first step, we have developed high-throughput live cell imaging in combination<br />
with RNAi screening to identify novel genes that function in the above cell division processes.<br />
Selected references<br />
Dultz, E., Zanin, E., Wurzenberger, C., Braun, M., Rabut, G., Sironi,<br />
L. & Ellenberg, J. (2008). Systematic kinetic analysis of mitotic disand<br />
reassembly of the nuclear pore in living cells. J. Cell Biol., 180,<br />
857-65<br />
Schuh, M. & Ellenberg, J. (2008). A new model for asymmetric<br />
spindle positioning in mouse oocytes. Curr. Biol., 18, 1986-92<br />
Schuh, M. & Ellenberg, J. (2007). Self-organization of MTOCs<br />
replaces centrosome function during acentrosomal spindle assembly<br />
in live mouse oocytes. Cell, 13, 8-98<br />
Neumann, B., Held, M., Liebel, U., Erfle, H., Rogers, P., Pepperkok,<br />
R. & Ellenberg, J. (2006). High-throughput RNAi screening by timelapse<br />
imaging of live human cells. Nat. Methods, 3, 385-90<br />
8