Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>EMBL</strong> Research at a Glance 2009<br />
Nicolas Le<br />
Novère<br />
PhD 1998, Pasteur Institute,<br />
Paris.<br />
Postdoctoral research at the<br />
University of Cambridge.<br />
Research fellow, CNRS,<br />
Paris.<br />
Group leader at <strong>EMBL</strong>-EBI<br />
since 2003.<br />
Computational systems neurobiology<br />
Previous and current research<br />
The Le Novère group’s research interests revolve around signal transduction in neurons, ranging<br />
from the molecular structure of membrane proteins involved in neurotransmission to modelling<br />
signalling pathways. In particular, we focus on the molecular and cellular basis of neuroadaptation<br />
in neurons of the basal ganglia. The supra-macromolecular structure of the postsynaptic membrane<br />
strongly influences signal transduction. Moreover, the whole structure is dynamic and<br />
evolves, for example, under the control of neuronal activity. By building detailed and realistic computational<br />
models, we try to understand how neurotransmitter-receptor movement and clustering,<br />
interactions between membrane and cytoplasmic proteins, and spatial location influence<br />
synaptic signalling. Downstream from the transduction machinery, we build quantitative models<br />
of the integration of signalling pathways known to mediate the effects of neurotransmitters, neuromodulators<br />
and drugs of abuse. We are particularly interested in understanding the processes<br />
of cooperativity, pathway switch and bistability.<br />
The group provides community services that facilitate research in computational systems biology.<br />
In particular, we are leading the efforts in encoding and annotating kinetic models in chemistry<br />
and cellular biology, including the creation of standard representations, the production of databases<br />
and software development. The Systems Biology Markup Language (SBML) is designed to<br />
facilitate the exchange of biological models between different types of software. As editors of the<br />
language, we also work on increasing its coverage, and are developing software to support SBML<br />
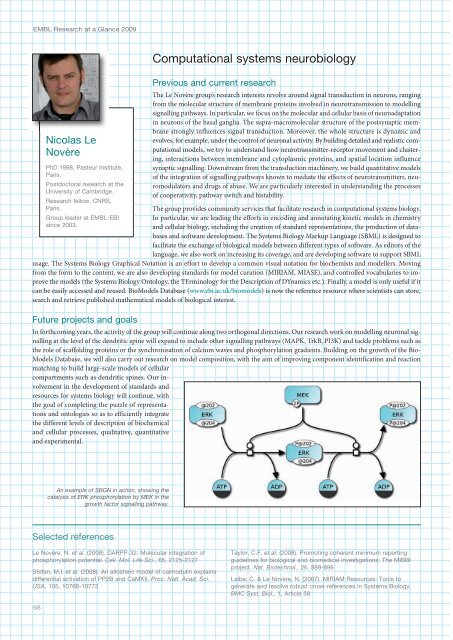
usage. The Systems Biology Graphical Notation is an effort to develop a common visual notation for biochemists and modellers. Moving<br />
from the form to the content, we are also developing standards for model curation (MIRIAM, MIASE), and controlled vocabularies to improve<br />
the models (the Systems Biology Ontology, the TErminology for the Description of DYnamics etc.). Finally, a model is only useful if it<br />
can be easily accessed and reused. BioModels Database (www.ebi.ac.uk/biomodels) is now the reference resource where scientists can store,<br />
search and retrieve published mathematical models of biological interest.<br />
Future projects and goals<br />
In forthcoming years, the activity of the group will continue along two orthogonal directions. Our research work on modelling neuronal signalling<br />
at the level of the dendritic spine will expand to include other signalling pathways (MAPK, TrkB, PI3K) and tackle problems such as<br />
the role of scaffolding proteins or the synchronisation of calcium waves and phosphorylation gradients. Building on the growth of the Bio-<br />
Models Database, we will also carry out research on model composition, with the aim of improving component identification and reaction<br />
matching to build large-scale models of cellular<br />
compartments such as dendritic spines. Our involvement<br />
in the development of standards and<br />
resources for systems biology will continue, with<br />
the goal of completing the puzzle of representations<br />
and ontologies so as to efficiently integrate<br />
the different levels of description of biochemical<br />
and cellular processes, qualitative, quantitative<br />
and experimental.<br />
An example of SBGN in action, showing the<br />
catalysis of ERK phosphorylation by MEK in the<br />
growth factor signalling pathway.<br />
Selected references<br />
Le Novère, N. et al. (2008). DARPP-32: Molecular integration of<br />
phosphorylation potential. Cell. Mol. Life Sci., 65, 2125-2127<br />
Stefan, M.I. et al. (2008). An allosteric model of calmodulin explains<br />
differential activation of PP2B and CaMKII. Proc. Natl. Acad. Sci.<br />
USA, 105, 10768-10773<br />
Taylor, C.F. et al. (2008). Promoting coherent minimum reporting<br />
guidelines for biological and biomedical investigations: The MIBBI<br />
project. Nat. Biotechnol., 26, 889-896<br />
Laibe, C. & Le Novère, N. (2007). MIRIAM Resources: Tools to<br />
generate and resolve robust cross-references in Systems Biology.<br />
BMC Syst. Biol., 1, Article 58<br />
68