You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>EMBL</strong> Research at a Glance 2009<br />
Christiane<br />
Schaffitzel<br />
PhD 2001, University of<br />
Zürich.<br />
Postdoctoral research at the<br />
Institute of Molecular Biology<br />
and Biophysics, ETH Zürich.<br />
Team leader at <strong>EMBL</strong><br />
<strong>Grenoble</strong> since 2007.<br />
Habilitation, ETH Zürich,<br />
2008.<br />
Cryo-EM of ribosomal complexes<br />
Previous and current research<br />
How exactly the folding of a nascent polypeptide chain into its native structure is achieved is one<br />
of the fundamental questions in biology. Synthesis and folding of proteins require concerted interactions<br />
of the translating ribosome with translation factors, processing enzymes, molecular<br />
chaperones and factors involved in the export of proteins. The functional and structural characterisation<br />
of such complexes is the aim of our team.<br />
We use cryo-EM and single particle analysis to study nascent chain co-translational folding, targeting<br />
and translocation. Structures of translating ribosomes in complex with the factors involved<br />
provide important mechanistic insight into the molecular mechanism of protein sorting.<br />
A prerequisite for any structural studies on translating ribosomes is to produce large amounts of homogenous,<br />
stable complexes. To this end, we adapted and decisively improved existing E. coli in<br />
vitro translation protocols which are optimised for protein expression for the production of ribosome<br />
nascent chain complexes. We have solved the structures of a translating ribosome-signal<br />
recognition particle (SRP) complex and of a non-translating ribosome without nascent chain in<br />
complex with SRP. Conformational changes of SRP which occur when SRP recognises a signal sequence<br />
on the nascent chain emerging from the ribosomal exit tunnel explain previous biochemical<br />
findings such as experimental crosslinks, salt-sensitivity of the 70S-SRP complex and a more<br />
than 100-fold improved affinity of SRP towards ribosomes displaying a nascent chain with a signal<br />
sequence. Furthermore, the structure of a translating ribosome in complex with the E. coli translocation machinery at 15 Å resolution reveals<br />
that both active and inactive translocon are composed of dimers of SecYEG. Cryo-EM analysis of a ribosome in complex with the molecular<br />
chaperone Trigger Factor suggests that Trigger Factor forms a hydrophobic arch over the ribosomal exit tunnel, thereby providing a folding interface<br />
for the nascent polypeptide and protection against other cytosolic proteins to prevent aggregation and degradation of nascent proteins.<br />
Our findings in aggregate support a view of the ribosomal exit tunnel as an anchor point for a multitude of factors involved in co-translational<br />
processes. A concerted action of these factors ensures correct processing, folding and targeting of nascent proteins.<br />
Future projects and goals<br />
In the future, we will continue analysing ribosomal complexes involved in co-translational processes to gain structural insight into all steps<br />
along the pathway of co-translational targeting and translocation.<br />
In collaboration with the Berger group (page 89), we study human macromolecular complexes involved in class II gene expression. We use<br />
EM to analyse the structure of these molecular machines, which we produce by means of advanced recombinant technologies.<br />
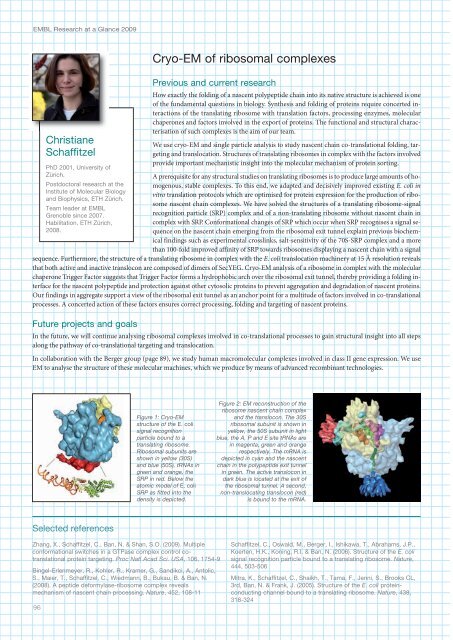
Figure 1: Cryo-EM<br />
structure of the E. coli<br />
signal recognition<br />
particle bound to a<br />
translating ribosome.<br />
Ribosomal subunits are<br />
shown in yellow (30S)<br />
and blue (50S), tRNAs in<br />
green and orange, the<br />
SRP in red. Below the<br />
atomic model of E. coli<br />
SRP as fitted into the<br />
density is depicted.<br />
Figure 2: EM reconstruction of the<br />
ribosome nascent chain complex<br />
and the translocon. The 30S<br />
ribosomal subunit is shown in<br />
yellow, the 50S subunit in light<br />
blue, the A, P and E site tRNAs are<br />
in magenta, green and orange<br />
respectively. The mRNA is<br />
depicted in cyan and the nascent<br />
chain in the polypeptide exit tunnel<br />
in green. The active translocon in<br />
dark blue is located at the exit of<br />
the ribosomal tunnel. A second,<br />
non-translocating translocon (red)<br />
is bound to the mRNA.<br />
Selected references<br />
Zhang, X., Schaffitzel, C., Ban, N. & Shan, S.O. (2009). Multiple<br />
conformational switches in a GTPase complex control cotranslational<br />
protein targeting. Proc Natl Acad Sci. USA, 106, 175-9<br />
Bingel-Erlenmeyer, R., Kohler, R., Kramer, G., Sandikci, A., Antolic,<br />
S., Maier, T., Schaffitzel, C., Wiedmann, B., Bukau, B. & Ban, N.<br />
(2008). A peptide deformylase-ribosome complex reveals<br />
mechanism of nascent chain processing. Nature, 52, 108-11<br />
96<br />
Schaffitzel, C., Oswald, M., Berger, I., Ishikawa, T., Abrahams, J.P.,<br />
Koerten, H.K., Koning, R.I. & Ban, N. (2006). Structure of the E. coli<br />
signal recognition particle bound to a translating ribosome. Nature,<br />
, 503-506<br />
Mitra, K., Schaffitzel, C., Shaikh, T., Tama, F., Jenni, S., Brooks CL,<br />
3rd, Ban, N. & Frank, J. (2005). Structure of the E. coli proteinconducting<br />
channel bound to a translating ribosome. Nature, 38,<br />
318-32