You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>EMBL</strong> Research at a Glance 2009<br />
Gene regulatory networks: Dissecting the logic<br />
Eileen Furlong<br />
PhD 1996, University College<br />
Dublin.<br />
Postdoctoral research at<br />
Stanford University.<br />
Group leader at <strong>EMBL</strong> since<br />
September 2002.<br />
Senior scientist and joint Unit<br />
Coordinator of Gene<br />
Expression Unit since 2009.<br />
Previous and current research<br />
Development is driven by the establishment of complex patterns of gene expression at precise<br />
times and spatial locations. Although a number of mechanisms fine-tune gene expression, it is<br />
initially established through the integration of signalling and transcriptional networks converging<br />
on enhancer elements, or cis-regulatory modules (CRMs). CRMs serve as regulatory circuits that<br />
integrate diverse transcriptional inputs leading to a specific spatio-temporal output of expression.<br />
Understanding how CRMs function is therefore central to understanding metazoan development<br />
and evolutionary change. Although there has been extensive progress in deciphering the function<br />
of individual CRMs, how these modules are integrated to regulate more global cis-regulatory networks<br />
remains a key challenge. Even in the extensively studied model organism, the Drosophila<br />
fruit fly, there are no predictive models for a transcriptional network leading to cell fate specification.<br />
The main aim of our research is to understand how gene regulatory networks control development<br />
and how network perturbations lead to specific phenotypes. To address this we integrate functional<br />
genomic, genetic and computational approaches to make predictive models of developmental<br />
progression. We are particularly interested in the topology and function of developmental<br />
networks at a global level.<br />
We use Drosophila mesoderm specification into different muscle primordia<br />
as a model system. The relative simplicity of the fly mesoderm,<br />
in addition to the number of essential and conserved<br />
transcription factors already identified, make it an ideal model to<br />
understand cell fate decisions at a systems level.<br />
Future projects and goals<br />
We have constructed a comprehensive cis-regulatory network during<br />
consecutive stages of development, elucidating the combinatorial<br />
binding and temporal occupancy of thousand of cis-regulatory<br />
modules, which we are currently using to understand differ aspects<br />
of gene expression. Our future work will involve extending this network<br />
to other species to study the evolution of cis-regulatory networks<br />
at a genome-wide scale. We are also generating genetic tools<br />
to allow us investigate changes in chromatin remodelling<br />
during cell fate decisions. Our ultimate<br />
goal is to use this systems-level approach<br />
to make predictive models of embryonic development<br />
and the effect of genetic perturbations.<br />
Working with Drosophila allows us to readily test<br />
all predictions on network perturbations during<br />
embryonic development.<br />
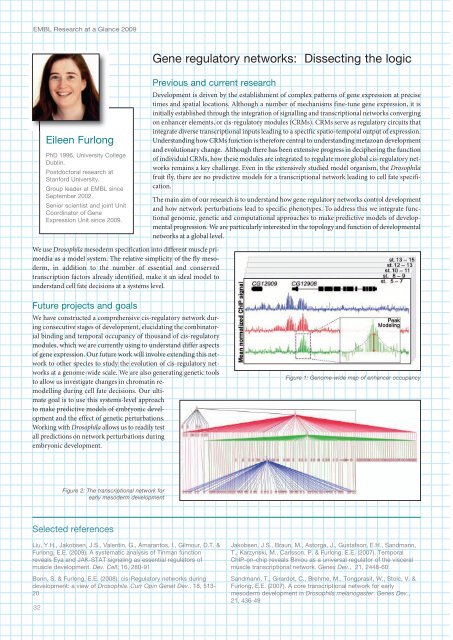
Figure 1: Genome-wide map of enhancer occupancy<br />
Figure 2: The transcriptional network for<br />
early mesoderm development<br />
Selected references<br />
Liu, Y.H., Jakobsen, J.S., Valentin, G., Amarantos, I., Gilmour, D.T. &<br />
Furlong, E.E. (2009). A systematic analysis of Tinman function<br />
reveals Eya and JAK-STAT signaling as essential regulators of<br />
muscle development. Dev. Cell, 16, 280-91<br />
Bonn, S. & Furlong, E.E. (2008). cis-Regulatory networks during<br />
development: a view of Drosophila. Curr Opin Genet Dev., 18, 513-<br />
20<br />
32<br />
Jakobsen, J.S., Braun, M., Astorga, J., Gustafson, E.H., Sandmann,<br />
T., Karzynski, M., Carlsson, P. & Furlong, E.E. (2007). Temporal<br />
ChIP-on-chip reveals Biniou as a universal regulator of the visceral<br />
muscle transcriptional network. Genes Dev., 21, 28-60<br />
Sandmann, T., Girardot, C., Brehme, M., Tongprasit, W., Stolc, V. &<br />
Furlong, E.E. (2007). A core transcriptional network for early<br />
mesoderm development in Drosophila melanogaster. Genes Dev.,<br />
21, 36-9