You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
<strong>EMBL</strong> Research at a Glance 2009<br />
Michael Knop<br />
PhD 1995, University of<br />
Stuttgart.<br />
Postdoctoral research at the<br />
MPI for Biochemistry, Munich<br />
and the Beatson Institute for<br />
Cancer Research, Glasgow.<br />
Group leader at the MPI for<br />
Biochemistry, Munich.<br />
Group leader at <strong>EMBL</strong> since<br />
2001.<br />
Systems biology of meiosis and mating in<br />
budding yeast<br />
Previous and current research<br />
Our group is interested in the various cellular processes that underlie the sexual cycle of budding<br />
yeast (mating and meiosis). In the past we have addressed the meiosis specific pathways that regulate<br />
spore morphogenesis with respect to spindle pole body function, membrane formation and<br />
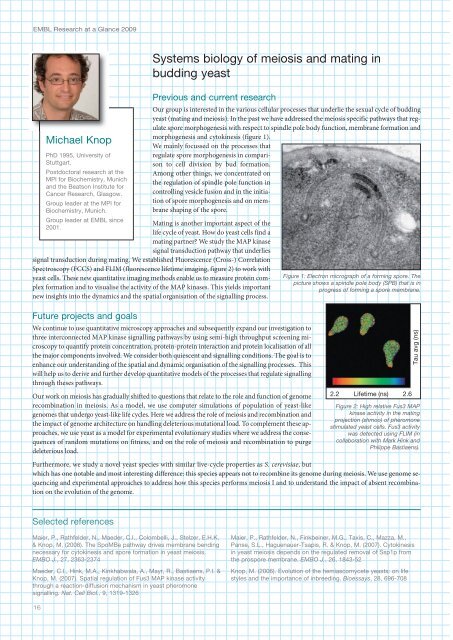
morphogenesis and cytokinesis (figure 1).<br />
We mainly focussed on the processes that<br />
regulate spore morphogenesis in comparison<br />
to cell division by bud formation.<br />
Among other things, we concentrated on<br />
the regulation of spindle pole function in<br />
controlling vesicle fusion and in the initiation<br />
of spore morphogenesis and on membrane<br />
shaping of the spore.<br />
Mating is another important aspect of the<br />
life cycle of yeast. How do yeast cells find a<br />
mating partner? We study the MAP kinase<br />
signal transduction pathway that underlies<br />
signal transduction during mating. We established Fluorescence (Cross-) Correlation<br />
Spectroscopy (FCCS) and FLIM (fluorescence lifetime imaging, figure 2) to work with<br />
yeast cells. These new quantitative imaging methods enable us to measure protein complex<br />
formation and to visualise the activity of the MAP kinases. This yields important<br />
new insights into the dynamics and the spatial organisation of the signalling process.<br />
Future projects and goals<br />
We continue to use quantitative microscopy approaches and subsequently expand our investigation to<br />
three interconnected MAP kinase signalling pathways by using semi-high throughput screening microscopy<br />
to quantify protein concentration, protein-protein interaction and protein localisation of all<br />
the major components involved. We consider both quiescent and signalling conditions. The goal is to<br />
enhance our understanding of the spatial and dynamic organisation of the signalling processes. This<br />
will help us to derive and further develop quantitative models of the processes that regulate signalling<br />
through theses pathways.<br />
Our work on meiosis has gradually shifted to questions that relate to the role and function of genome<br />
recombination in meiosis. As a model, we use computer simulations of population of yeast-like<br />
genomes that undergo yeast-like life cycles. Here we address the role of meiosis and recombination and<br />
the impact of genome architecture on handling deleterious mutational load. To complement these approaches,<br />
we use yeast as a model for experimental evolutionary studies where we address the consequences<br />
of random mutations on fitness, and on the role of meiosis and recombination to purge<br />
deleterious load.<br />
Figure 1: Electron micrograph of a forming spore. The<br />
picture shows a spindle pole body (SPB) that is in<br />
progress of forming a spore membrane.<br />
2.2 Lifetime (ns) 2.6<br />
Furthermore, we study a novel yeast species with similar live-cycle properties as S. cerevisiae, but<br />
which has one notable and most interesting difference: this species appears not to recombine its genome during meiosis. We use genome sequencing<br />
and experimental approaches to address how this species performs meiosis I and to understand the impact of absent recombination<br />
on the evolution of the genome.<br />
Tau avg (ns)<br />
Figure 2: High relative Fus3 MAP<br />
kinase activity in the mating<br />
projection (shmoo) of pheromone<br />
stimulated yeast cells. Fus3 activity<br />
was detected using FLIM (in<br />
collaboration with Mark Hink and<br />
Philippe Bastiaens).<br />
Selected references<br />
Maier, P., Rathfelder, N., Maeder, C.I., Colombelli, J., Stelzer, E.H.K.<br />
& Knop, M. (2008). The SpoMBe pathway drives membrane bending<br />
necessary for cytokinesis and spore formation in yeast meiosis.<br />
EMBO J., 27, 2363-237<br />
Maeder, C.I., Hink, M.A., Kinkhabwala, A., Mayr, R., Bastiaens, P.I. &<br />
Knop, M. (2007). Spatial regulation of Fus3 MAP kinase activity<br />
through a reaction-diffusion mechanism in yeast pheromone<br />
signalling. Nat. Cell Biol., 9, 1319-1326<br />
Maier, P., Rathfelder, N., Finkbeiner, M.G., Taxis, C., Mazza, M.,<br />
Panse, S.L., Haguenauer-Tsapis, R. & Knop, M. (2007). Cytokinesis<br />
in yeast meiosis depends on the regulated removal of Ssp1p from<br />
the prospore membrane. EMBO J., 26, 183-52<br />
Knop, M. (2006). Evolution of the hemiascomycete yeasts: on life<br />
styles and the importance of inbreeding. Bioessays, 28, 696-708<br />
16